Search Count: 12
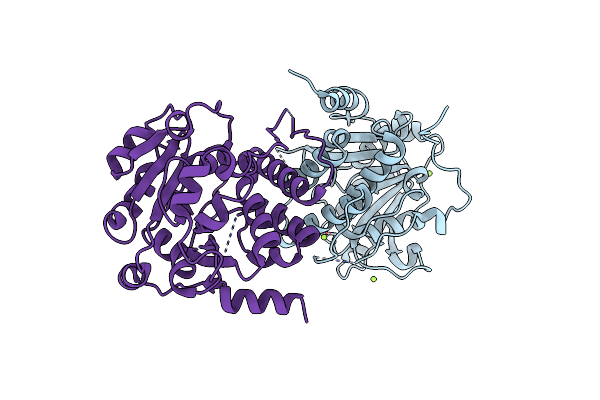 |
Crystal Structure Of Mycobacterium Hassiacum Glucosyl-3-Phosphoglycerate Synthase At Ph 8.5 - Apo Form
Organism: Mycolicibacterium hassiacum (strain dsm 44199 / cip 105218 / jcm 12690 / 3849)
Method: X-RAY DIFFRACTION Resolution:1.43 Å Release Date: 2023-04-19 Classification: TRANSFERASE Ligands: CL, MG |
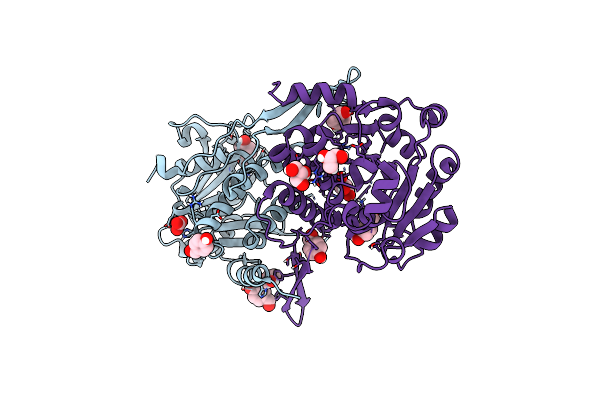 |
Crystal Structure Of Mycobacterium Hassiacum Glucosyl-3-Phosphoglycerate Synthase At Ph 7.1 In Complex With 4-Hydroxybenzaldehyde
Organism: Mycolicibacterium hassiacum (strain dsm 44199 / cip 105218 / jcm 12690 / 3849)
Method: X-RAY DIFFRACTION Resolution:1.27 Å Release Date: 2023-03-01 Classification: TRANSFERASE Ligands: HBA, BCT, GOL, MLT |
 |
Crystal Structure Of Mycobacterium Hassiacum Glucosyl-3-Phosphoglycerate Synthase At Ph 5.5 In Complex With Udp-Glucose
Organism: Mycolicibacterium hassiacum (strain dsm 44199 / cip 105218 / jcm 12690 / 3849)
Method: X-RAY DIFFRACTION Resolution:2.05 Å Release Date: 2023-02-22 Classification: TRANSFERASE Ligands: UPG, MG, CL, BCT |
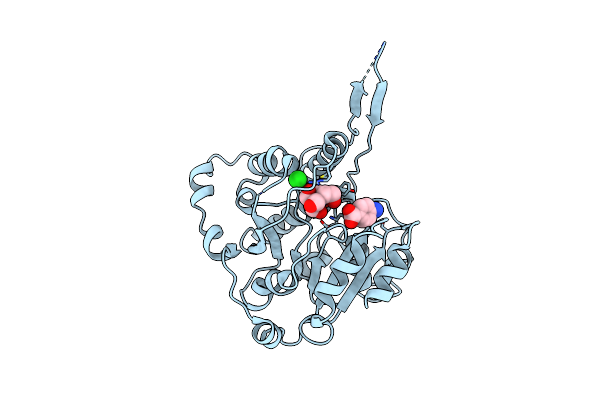 |
Crystal Structure Of Mycobacterium Hassiacum Glucosyl-3-Phosphoglycerate Synthase At Ph 5.5 In Complex With 4-Aminobenzoic Acid
Organism: Mycolicibacterium hassiacum (strain dsm 44199 / cip 105218 / jcm 12690 / 3849)
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2023-02-15 Classification: TRANSFERASE Ligands: PAB, BTB, CL |
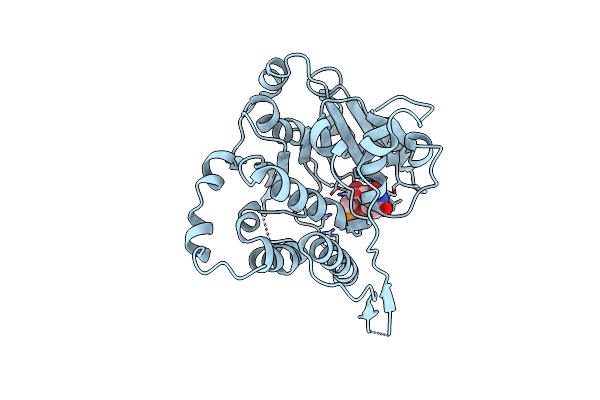 |
Crystal Structure Of Mycobacterium Hassiacum Glucosyl-3-Phosphoglycerate Synthase At Ph 5.5 In Complex With Udp
Organism: Mycolicibacterium hassiacum dsm 44199
Method: X-RAY DIFFRACTION Resolution:1.97 Å Release Date: 2023-02-15 Classification: TRANSFERASE Ligands: CL, UDP |
 |
Crystal Structure Of Mycobacterium Hassiacum Glucosyl-3-Phosphoglycerate Synthase At Ph 5.5 - Apo Form
Organism: Mycolicibacterium hassiacum dsm 44199
Method: X-RAY DIFFRACTION Resolution:2.13 Å Release Date: 2023-01-25 Classification: TRANSFERASE Ligands: CL, BGC, MLI |
 |
Crystal Structure Of Mycobacterium Hassiacum Glucosyl-3-Phosphoglycerate Synthase At Ph 7.1 - Apo Form
Organism: Mycolicibacterium hassiacum (strain dsm 44199 / cip 105218 / jcm 12690 / 3849)
Method: X-RAY DIFFRACTION Resolution:1.22 Å Release Date: 2023-01-18 Classification: TRANSFERASE Ligands: LMR, MLT |
 |
Crystal Structure Of Mycobacterium Hassiacum Glucosyl-3-Phosphoglycerate Synthase At Ph 8.5 In Complex With Ump And Magnesium
Organism: Mycolicibacterium hassiacum dsm 44199
Method: X-RAY DIFFRACTION Release Date: 2023-01-18 Classification: TRANSFERASE Ligands: MG, U5P, CL |
 |
Crystal Structure Of Mycobacterium Hassiacum Glucosyl-3-Phosphoglycerate Synthase At Ph 7.2 In Complex With Udp
Organism: Mycolicibacterium hassiacum dsm 44199
Method: X-RAY DIFFRACTION Resolution:1.23 Å Release Date: 2022-12-28 Classification: TRANSFERASE Ligands: UDP |
 |
Crystal Structure Of Mycobacterium Hassiacum Glucosyl-3-Phosphoglycerate Synthase At Ph 8.5 In Complex With Udp
Organism: Mycolicibacterium hassiacum dsm 44199
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2022-12-28 Classification: TRANSFERASE Ligands: MG, UDP, CL |
 |
Crystal Structure Of Mycobacterium Hassiacum Glucosyl-3-Phosphoglycerate Synthase At Ph 7.2 - Apo Form
Organism: Mycolicibacterium hassiacum (strain dsm 44199 / cip 105218 / jcm 12690 / 3849)
Method: X-RAY DIFFRACTION Resolution:1.11 Å Release Date: 2022-12-14 Classification: TRANSFERASE Ligands: TLA |
 |
Human Procarboxypeptidase B: Three-Dimensional Structure And Implications For Thrombin-Activatable Fibrinolysis Inhibitor (Tafi)
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2002-06-05 Classification: HYDROLASE Ligands: ZN, CIT |

