Search Count: 18
 |
Organism: Vairimorpha necatrix
Method: ELECTRON MICROSCOPY Release Date: 2022-11-23 Classification: HYDROLASE |
 |
Organism: Enterococcus faecalis, Vicugna pacos
Method: ELECTRON MICROSCOPY Release Date: 2022-05-18 Classification: MEMBRANE PROTEIN |
 |
Organism: vibrio cholerae o1 biovar el tor str. n16961
Method: ELECTRON MICROSCOPY Release Date: 2022-02-23 Classification: TOXIN |
 |
Organism: Paranosema locustae
Method: ELECTRON MICROSCOPY Release Date: 2020-11-04 Classification: RIBOSOME Ligands: MG, ZN, AMP |
 |
Evolutionary Compaction And Adaptation Visualized By The Structure Of The Dormant Microsporidian Ribosome
Organism: Vairimorpha necatrix
Method: ELECTRON MICROSCOPY Release Date: 2019-07-10 Classification: RIBOSOME Ligands: MG, ZN |
 |
Conformational Switches Control Early Maturation Of The Eukaryotic Small Ribosomal Subunit
Organism: Saccharomyces cerevisiae by4741
Method: ELECTRON MICROSCOPY Release Date: 2019-06-19 Classification: RIBOSOME |
 |
Organism: Saccharomyces cerevisiae by4741
Method: ELECTRON MICROSCOPY Release Date: 2018-03-14 Classification: RIBOSOME Ligands: ZN |
 |
Organism: Saccharomyces cerevisiae by4741
Method: ELECTRON MICROSCOPY Release Date: 2018-03-14 Classification: RIBOSOME Ligands: ZN |
 |
Organism: Saccharomyces cerevisiae by4741
Method: ELECTRON MICROSCOPY Release Date: 2017-09-27 Classification: RIBOSOME Ligands: ZN |
 |
Organism: Saccharomyces cerevisiae s288c, Saccharomyces cerevisiae s288c
Method: ELECTRON MICROSCOPY Release Date: 2016-12-21 Classification: TRANSLATION |
 |
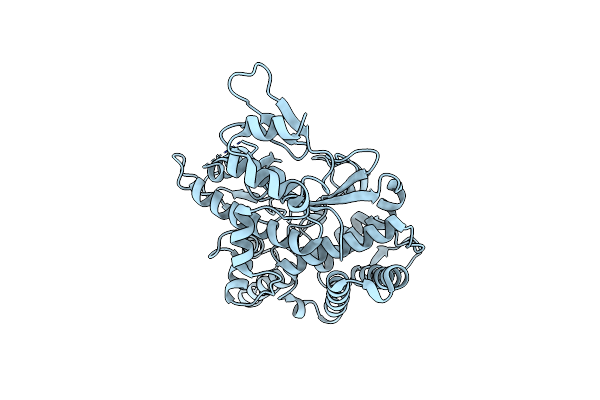
Crystal Structure Of The Complex Between Prokaryotic Ubiquitin-Like Protein Pup And Its Ligase Pafa
Organism: Corynebacterium glutamicum
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2013-05-01 Classification: LIGASE Ligands: ATP, MG |
 |
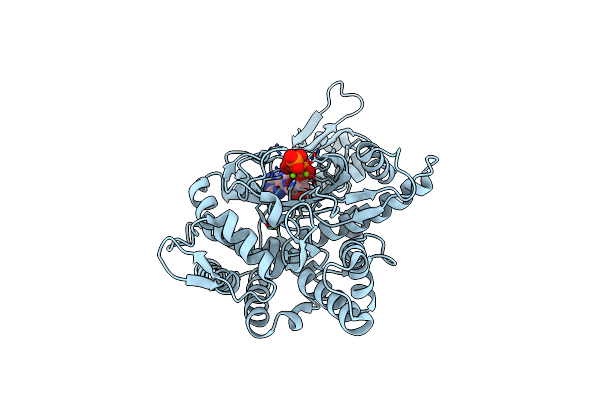
Structure Of The Deamidase-Depupylase Dop Of The Prokaryotic Ubiquitin-Like Modification Pathway
Organism: Acidothermus cellulolyticus
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2012-09-12 Classification: HYDROLASE |
 |
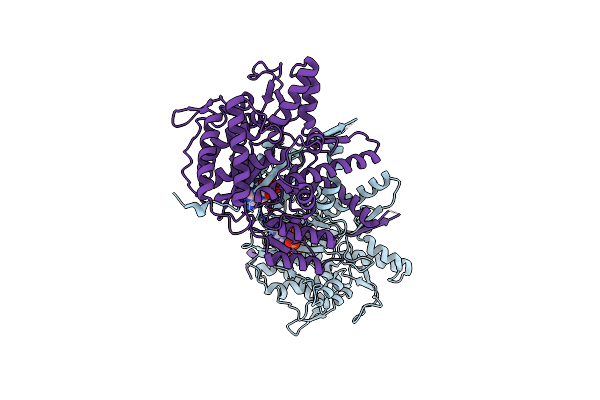
Structure Of The Deamidase-Depupylase Dop Of The Prokaryotic Ubiquitin-Like Modification Pathway In Complex With Atp
Organism: Acidothermus cellulolyticus
Method: X-RAY DIFFRACTION Resolution:2.85 Å Release Date: 2012-09-12 Classification: HYDROLASE Ligands: ATP, MG |
 |
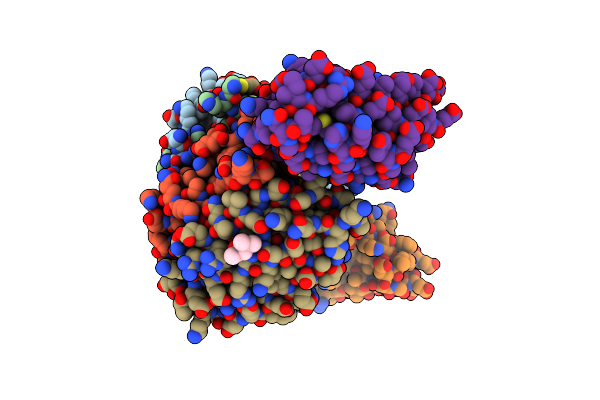
Structure Of The Pup Ligase Pafa Of The Prokaryotic Ubiquitin-Like Modification Pathway In Complex With Adp
Organism: Corynebacterium glutamicum
Method: X-RAY DIFFRACTION Resolution:2.16 Å Release Date: 2012-09-12 Classification: LIGASE Ligands: ADP, MG |
 |
Organism: Homo sapiens, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2011-12-28 Classification: HYDROLASE/INHIBITOR Ligands: SO4, EDO |
 |
Organism: Homo sapiens, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2011-12-21 Classification: HYDROLASE/PROTEIN BINDING Ligands: MPD, MRD |
 |
Organism: Homo sapiens, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2011-12-14 Classification: HYDROLASE/PROTEIN BINDING Ligands: MRD, MPD |
 |
Organism: Homo sapiens, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2011-12-14 Classification: HYDROLASE/PROTEIN BINDING Ligands: MRD |

