Search Count: 22
 |
E. Coli Dna-Directed Rna Polymerase Transcription Elongation Complex Bound To The Unnatural Dz-Ptp Base Pair In The Active Site
Organism: Escherichia coli, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2024-01-24 Classification: TRANSCRIPTION Ligands: ZN, MG, S9F |
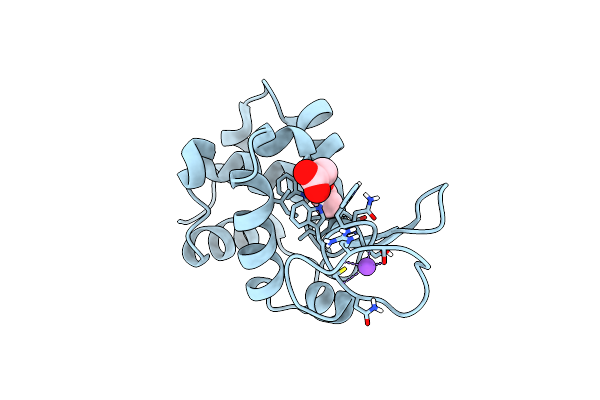 |
Organism: Gallus gallus
Method: X-RAY DIFFRACTION Resolution:1.57 Å Release Date: 2022-06-22 Classification: HYDROLASE |
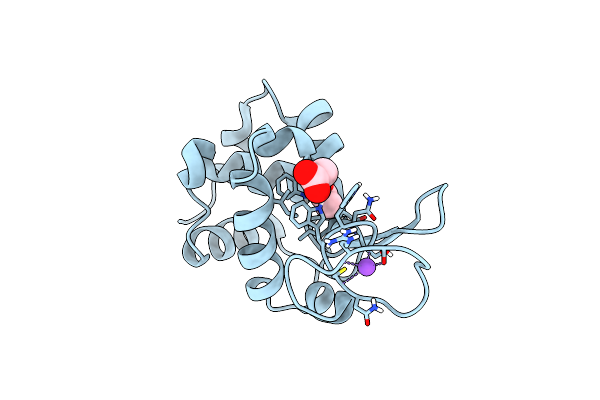 |
Organism: Gallus gallus
Method: X-RAY DIFFRACTION Resolution:1.57 Å Release Date: 2022-06-22 Classification: HYDROLASE |
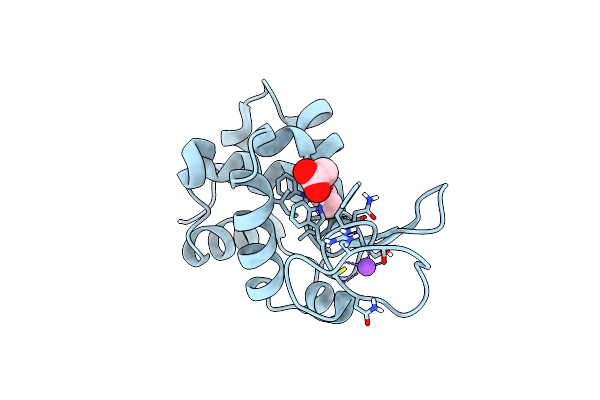 |
Organism: Gallus gallus
Method: X-RAY DIFFRACTION Resolution:1.57 Å Release Date: 2022-06-22 Classification: HYDROLASE |
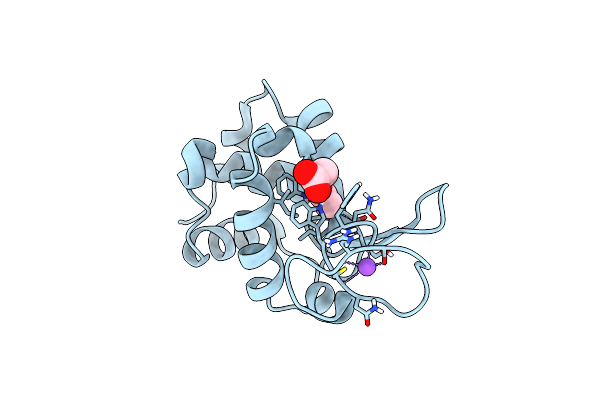 |
Organism: Gallus gallus
Method: X-RAY DIFFRACTION Resolution:1.57 Å Release Date: 2022-06-22 Classification: HYDROLASE |
 |
Organism: Gallus gallus
Method: X-RAY DIFFRACTION Resolution:1.57 Å Release Date: 2022-06-22 Classification: HYDROLASE |
 |
Organism: Gallus gallus
Method: X-RAY DIFFRACTION Resolution:1.57 Å Release Date: 2022-06-22 Classification: HYDROLASE |
 |
Organism: Gallus gallus
Method: X-RAY DIFFRACTION Resolution:1.57 Å Release Date: 2022-06-22 Classification: HYDROLASE |
 |
Organism: Gallus gallus
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2022-06-22 Classification: HYDROLASE |
 |
Organism: Gallus gallus
Method: X-RAY DIFFRACTION Resolution:1.57 Å Release Date: 2022-06-22 Classification: HYDROLASE |
 |
Organism: Gallus gallus
Method: X-RAY DIFFRACTION Resolution:1.57 Å Release Date: 2022-06-22 Classification: HYDROLASE |
 |
Laser Off Temperature-Jump Xfel Structure Of Lysozyme Bound To N,N'-Diacetylchitobiose
Organism: Gallus gallus
Method: X-RAY DIFFRACTION Resolution:1.51 Å Release Date: 2022-06-22 Classification: HYDROLASE |
 |
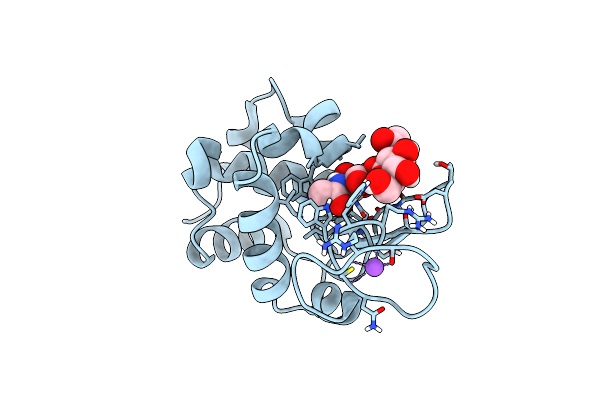
20Ns Temperature-Jump (Light) Xfel Structure Of Lysozyme Bound To N,N'-Diacetylchitobiose
Organism: Gallus gallus
Method: X-RAY DIFFRACTION Resolution:1.48 Å Release Date: 2022-06-22 Classification: HYDROLASE |
 |
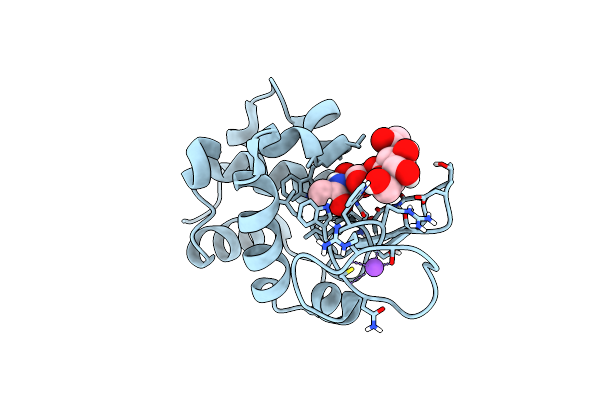
20Ns Temperature-Jump (Dark1) Xfel Structure Of Lysozyme Bound To N,N'-Diacetylchitobiose
Organism: Gallus gallus
Method: X-RAY DIFFRACTION Resolution:1.48 Å Release Date: 2022-06-22 Classification: HYDROLASE |
 |
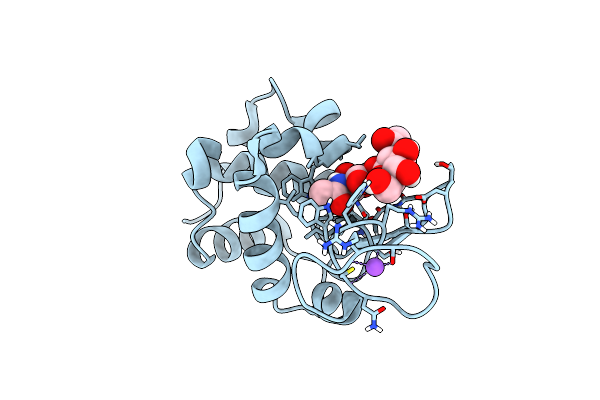
20Ns Temperature-Jump (Dark2) Xfel Structure Of Lysozyme Bound To N,N'-Diacetylchitobiose
Organism: Gallus gallus
Method: X-RAY DIFFRACTION Resolution:1.48 Å Release Date: 2022-06-22 Classification: HYDROLASE |
 |
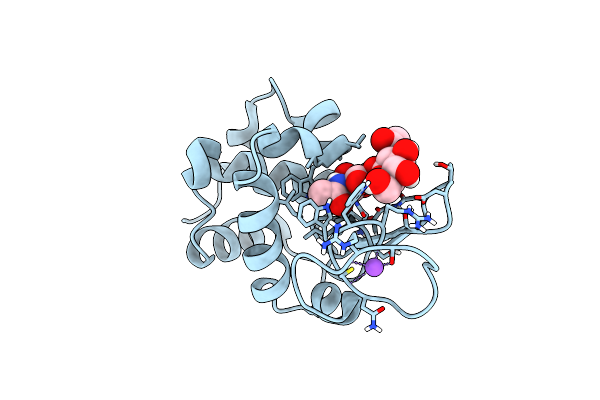
200Us Temperature-Jump (Light) Xfel Structure Of Lysozyme Bound To N,N'-Diacetylchitobiose
Organism: Gallus gallus
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2022-06-22 Classification: HYDROLASE |
 |
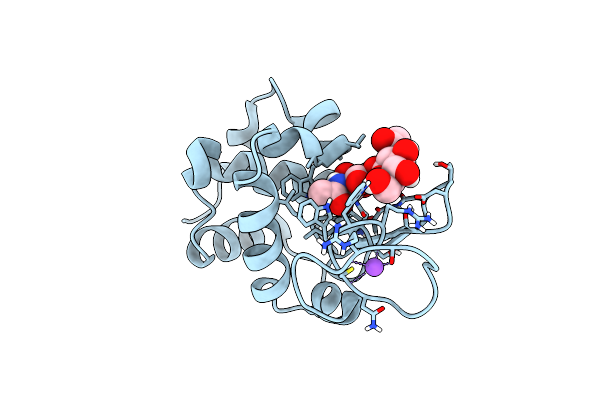
200Us Temperature-Jump (Dark1) Xfel Structure Of Lysozyme Bound To N,N'-Diacetylchitobiose
Organism: Gallus gallus
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2022-06-22 Classification: HYDROLASE |
 |
200Us Temperature-Jump (Dark2) Xfel Structure Of Lysozyme Bound To N,N'-Diacetylchitobiose
Organism: Gallus gallus
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2022-06-22 Classification: HYDROLASE |
 |
Xfel Structure Of Influenza A M2 Wild Type Tm Domain At High Ph In The Lipidic Cubic Phase At Room Temperature
Organism: Influenza a virus (a/hickox/1940(h1n1))
Method: X-RAY DIFFRACTION Resolution:1.40 Å Release Date: 2017-08-23 Classification: MEMBRANE PROTEIN Ligands: CL, CA |
 |
Xfel Structure Of Influenza A M2 Wild Type Tm Domain At Intermediate Ph In The Lipidic Cubic Phase At Room Temperature
Organism: Influenza a virus (a/hickox/1940(h1n1))
Method: X-RAY DIFFRACTION Resolution:1.45 Å Release Date: 2017-08-23 Classification: PROTON TRANSPORT Ligands: CA, CL |

