Search Count: 52
 |
Organism: Achromobacter xylosoxidans nbrc 15126 = atcc 27061
Method: X-RAY DIFFRACTION Resolution:2.11 Å Release Date: 2024-01-10 Classification: DNA BINDING PROTEIN |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2020-11-18 Classification: HYDROLASE Ligands: MN, FE, PO4 |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2018-11-21 Classification: HYDROLASE Ligands: PO4, FE, MN |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2018-11-21 Classification: HYDROLASE Ligands: FE, MN, PO4 |
 |
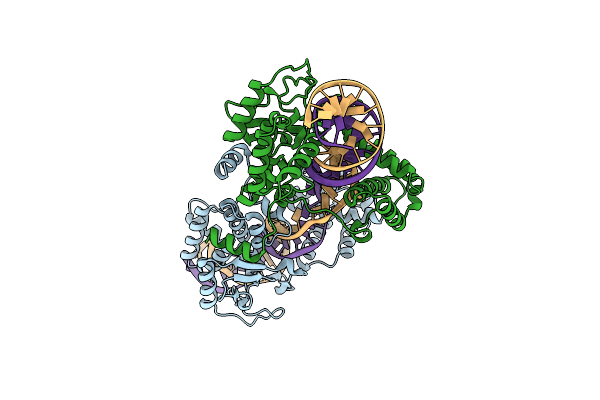
Structure Of The Tn1549 Transposon Integrase (Aa 82-397, Y379F) In Complex With Transposon Right End Dna
Organism: Enterococcus faecalis, Clostridioides difficile
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2018-04-04 Classification: RECOMBINATION |
 |
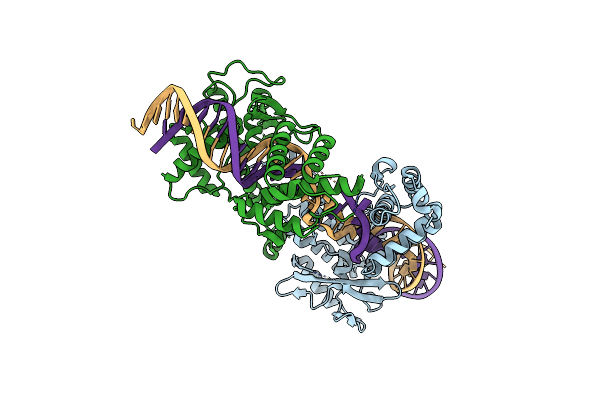
Structure Of The Tn1549 Transposon Integrase (Aa 82-397, R225K) In Complex With Circular Intermediate Dna (Ci5-Dna)
Organism: Enterococcus faecalis, Clostridioides difficile
Method: X-RAY DIFFRACTION Resolution:2.79 Å Release Date: 2018-04-04 Classification: RECOMBINATION |
 |
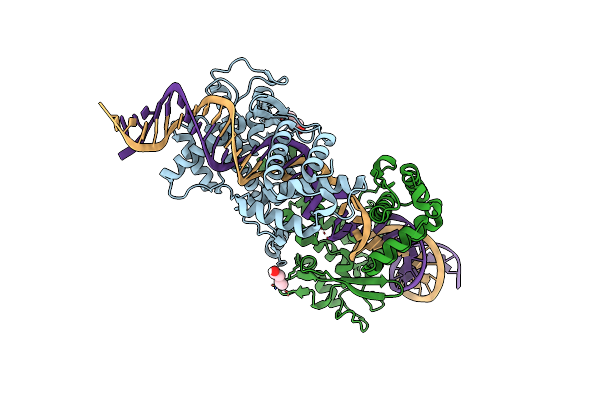
Structure Of The Tn1549 Transposon Integrase (Aa 82-397) In Complex With Circular Intermediate Dna (Ci5-Dna)
Organism: Enterococcus faecalis, Clostridioides difficile
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2018-04-04 Classification: RECOMBINATION |
 |
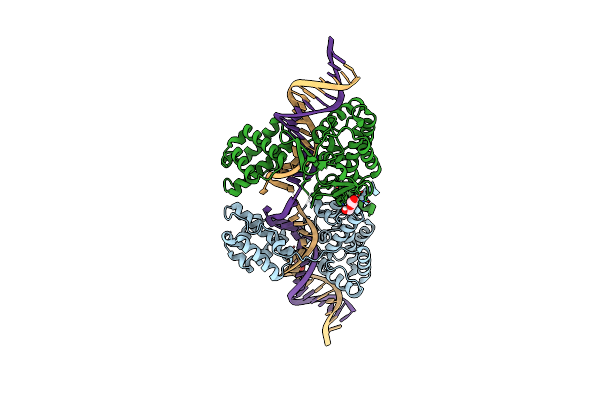
Structure Of The Tn1549 Transposon Integrase (Aa 82-397, R225K) In Complex With A Circular Intermediate Dna (Ci6A-Dna)
Organism: Enterococcus faecalis
Method: X-RAY DIFFRACTION Resolution:2.67 Å Release Date: 2018-04-04 Classification: RECOMBINATION Ligands: PEG |
 |
Structure Of The Tn1549 Transposon Integrase (Aa 82-397, R225K) In Complex With A Circular Intermediate Dna (Ci6B-Dna)
Organism: Enterococcus faecalis
Method: X-RAY DIFFRACTION Resolution:2.67 Å Release Date: 2018-04-04 Classification: RECOMBINATION Ligands: PEG |
 |
Crystal Structure Of The Is608 Transposase In Complex With Left End 29-Mer Dna Hairpin And A 6-Mer Dna Representing The Intact Target Site: Pre-Cleavage Target Capture Complex
Organism: Helicobacter pylori
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2018-03-28 Classification: DNA BINDING PROTEIN Ligands: CA |
 |
Organism: Escherichia coli s88, Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.51 Å Release Date: 2017-10-04 Classification: RNA Ligands: SO4 |
 |
Crystal Structure Of Xerh Site-Specific Recombinase Bound To Palindromic Difh Substrate: Post-Cleavage Complex
Organism: Helicobacter pylori (strain atcc 700392 / 26695), Helicobacter pylori 26695
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2016-12-28 Classification: RECOMBINATION Ligands: GOL, CL |
 |
Crystal Structure Of Xerh Site-Specific Recombinase Bound To Difh Substrate: Pre-Cleavage Complex
Organism: Helicobacter pylori (strain atcc 700392 / 26695), Helicobacter pylori 26695
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2016-12-28 Classification: CELL CYCLE Ligands: GOL, EDO, CL, PEG |
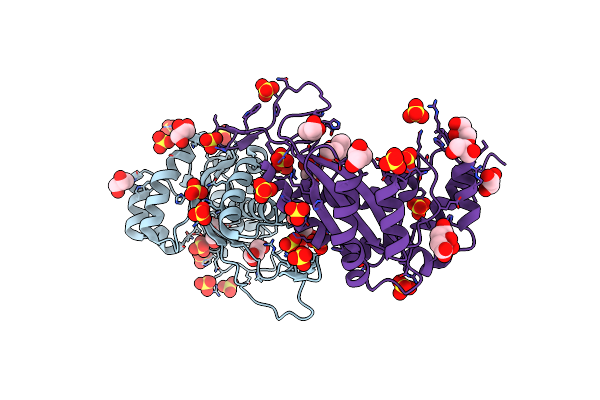 |
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.40 Å Release Date: 2016-03-30 Classification: HYDROLASE Ligands: SO4, GOL, CIT, EPE |
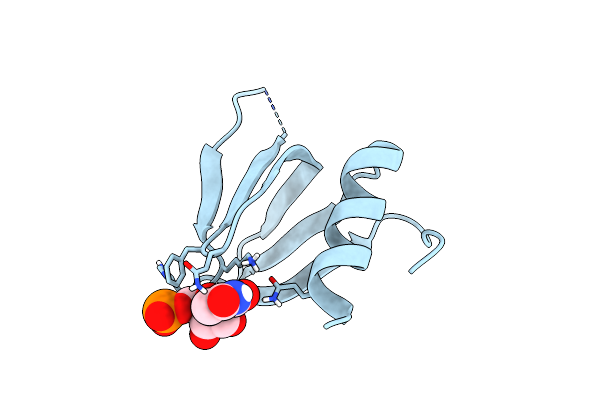 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:0.97 Å Release Date: 2014-11-19 Classification: RNA BINDING PROTEIN Ligands: U5P |
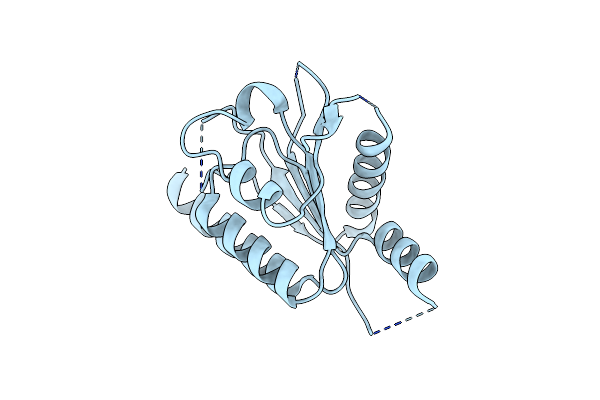 |
Crystal Structure Of The C-Terminal Domain Of Drosophila Melanogaster Zucchini
Organism: Drosophila melanogaster
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2012-11-07 Classification: HYDROLASE |
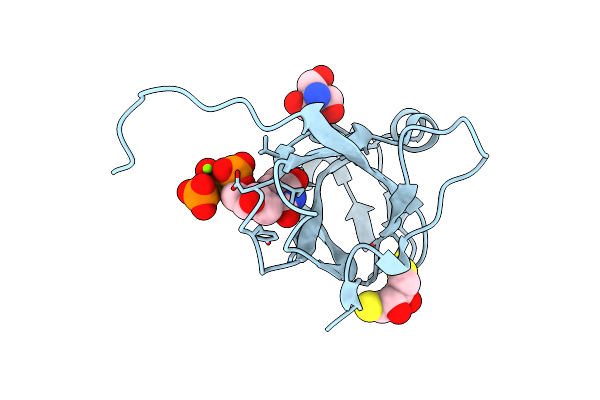 |
Organism: Mason-pfizer monkey virus
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2011-10-12 Classification: HYDROLASE Ligands: DUP, MG, TRS, DTT |
 |
Organism: Mason-pfizer monkey virus
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2011-10-12 Classification: HYDROLASE Ligands: MG, DUP, TRS |
 |
Crystal Structure Of M-Pmv Dutpase With A Mixed Population Of Substrate (Dupnpp) And Post-Inversion Product (Dump) In The Active Sites
Organism: Mason-pfizer monkey virus
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2011-10-12 Classification: HYDROLASE Ligands: MG, TRS, DUP, UMP |
 |
Crystal Structure Of M-Pmv Dutpase With A Mixed Population Of Substrate (Dupnpp) And Post-Inversion Product (Dump) In The Active Sites
Organism: Mason-pfizer monkey virus
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2011-10-12 Classification: HYDROLASE Ligands: UMP, DUP, MG |

