Search Count: 25
 |
Organism: Escherichia coli k-12
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2019-01-16 Classification: LYASE Ligands: FE, SF4, PEG, MG, GOL |
 |
Organism: Bacillus subtilis (strain 168)
Method: X-RAY DIFFRACTION Resolution:1.29 Å Release Date: 2018-10-24 Classification: OXIDOREDUCTASE Ligands: SAM, SF4, FES, MG |
 |
Organism: Methanocaldococcus jannaschii
Method: X-RAY DIFFRACTION Resolution:1.64 Å Release Date: 2018-06-13 Classification: METAL BINDING PROTEIN Ligands: SF4, GOL, PEG, FS0 |
 |
Organism: Bacillus subtilis
Method: X-RAY DIFFRACTION Resolution:2.55 Å Release Date: 2017-01-18 Classification: LYASE Ligands: SF4, MET, PEG |
 |
Crystal Structure Of Quee From Bacillus Subtilis With 6-Carboxypterin-5'-Deoxyadenosyl Ester Bound
Organism: Bacillus subtilis
Method: X-RAY DIFFRACTION Resolution:2.41 Å Release Date: 2017-01-18 Classification: LYASE Ligands: SF4, 7C5, MET |
 |
Organism: Bacillus subtilis (strain 168)
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2016-09-28 Classification: OXIDOREDUCTASE Ligands: SF4, B12, GOL, PO4, CL |
 |
Organism: Bacillus subtilis (strain 168), Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2016-09-28 Classification: oxidoreductase/RNA Ligands: SF4, B12, GOL, PO4 |
 |
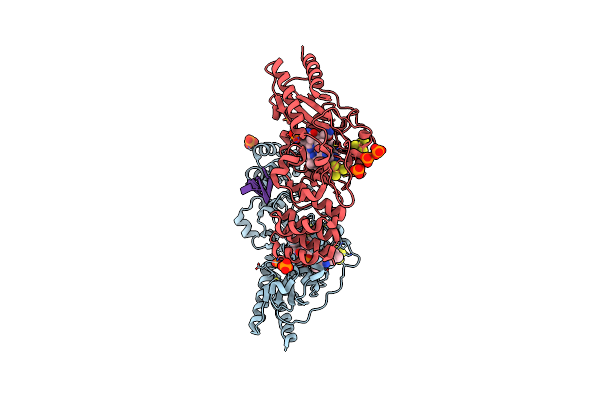
Crystal Structure Of Epoxyqueuosine Reductase With A Trna-Tyr Epoxyqueuosine-Modified Trna Stem Loop
Organism: Bacillus subtilis (strain 168), Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.65 Å Release Date: 2016-09-28 Classification: oxidoreductase/RNA Ligands: SF4, B12, PO4, GOL |
 |
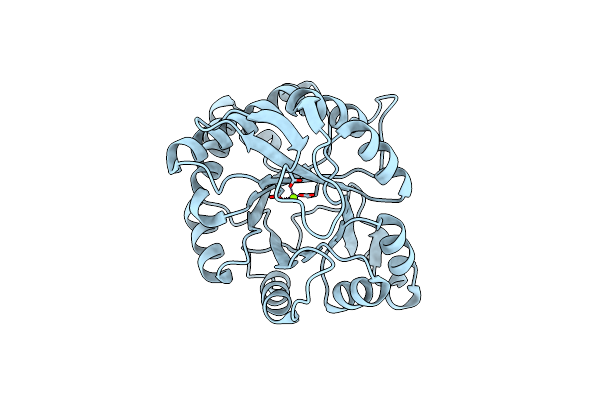
Structure Of Epoxyqueuosine Reductase From Bacillus Subtilis With The Asp134 Catalytic Loop Swung Out Of The Active Site.
Organism: Bacillus subtilis (strain 168), Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.65 Å Release Date: 2016-09-28 Classification: OXIDOREDUCTASE/RNA Ligands: SF4, B12, PO4 |
 |
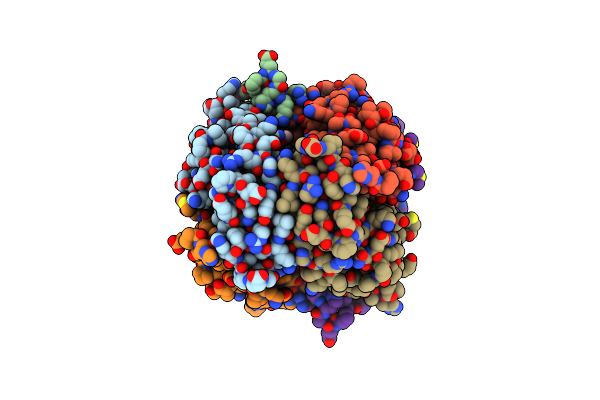
Structure Of Phospholipase D Beta1B1I From Sicarius Terrosus Venom At 2.14 A Resolution
Organism: Sicarius terrosus
Method: X-RAY DIFFRACTION Resolution:2.14 Å Release Date: 2015-03-18 Classification: LYASE Ligands: MG |
 |
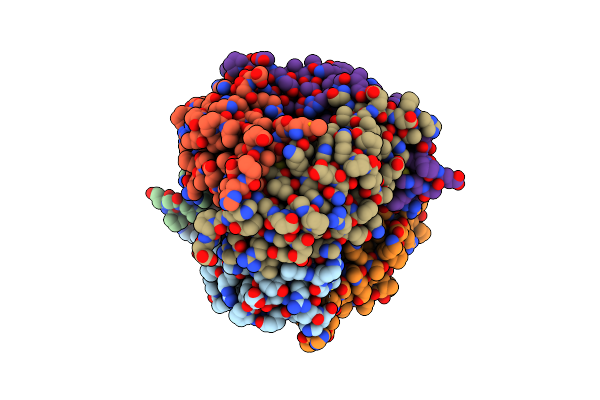
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2014-07-16 Classification: LYASE Ligands: ZN, ZSP, ACT |
 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.05 Å Release Date: 2014-07-16 Classification: LYASE Ligands: ZN, 2K8 |
 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.99 Å Release Date: 2014-07-16 Classification: LYASE Ligands: ZN, FMT |
 |
Crystal Structure Of Quee From Burkholderia Multivorans In Complex With Adomet And 6-Carboxypterin
Organism: Burkholderia multivorans
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2013-12-25 Classification: LYASE Ligands: SF4, SAM, HHS |
 |
Crystal Structure Of Quee From Burkholderia Multivorans In Complex With Adomet And 6-Carboxy-5,6,7,8-Tetrahydropterin
Organism: Burkholderia multivorans
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2013-12-25 Classification: LYASE Ligands: SF4, SAM, 2K8, NA, PO4 |
 |
Crystal Structure Of Quee From Burkholderia Multivorans In Complex With Adomet, 6-Carboxy-5,6,7,8-Tetrahydropterin, And Mg2+
Organism: Burkholderia multivorans
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2013-12-25 Classification: LYASE Ligands: SF4, SAM, 2K8, MG, PO4, NA |
 |
Crystal Structure Of Quee From Burkholderia Multivorans In Complex With Adomet, 6-Carboxy-5,6,7,8-Tetrahydropterin, And Manganese(Ii)
Organism: Burkholderia multivorans
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2013-12-25 Classification: LYASE Ligands: SF4, SAM, 2K8, MN |
 |
Crystal Structure Of Quee From Burkholderia Multivorans In Complex With Adomet, 7-Carboxy-7-Deazaguanine, And Mg2+
Organism: Burkholderia multivorans
Method: X-RAY DIFFRACTION Resolution:1.91 Å Release Date: 2013-12-25 Classification: LYASE Ligands: SF4, SAM, 2KA, MG, PO4, NA |
 |
Structural Analysis Of A Class I Preq1 Riboswitch Aptamer In The Metabolite-Bound State
Method: X-RAY DIFFRACTION
Resolution:2.75 Å Release Date: 2011-05-18 Classification: RNA Ligands: PRF, SO4 |
 |
Structural Analysis Of A Class I Preq1 Riboswitch Aptamer In The Metabolite-Free State.
Method: X-RAY DIFFRACTION
Resolution:2.85 Å Release Date: 2011-05-18 Classification: RNA Ligands: SO4, MG |

