Search Count: 8
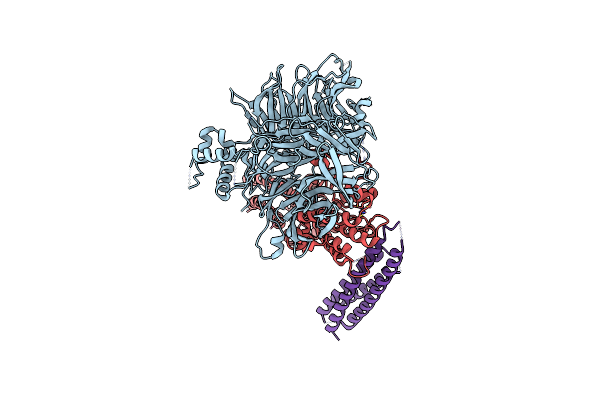 |
Cryo-Em Structure Of Rcmv-E E27 Bound To Human Ddb1 (Deltabpb) And Rat Stat2 Ccd
Organism: Homo sapiens, Murid betaherpesvirus 8, Rattus norvegicus
Method: ELECTRON MICROSCOPY Release Date: 2022-11-09 Classification: VIRUS Ligands: ZN |
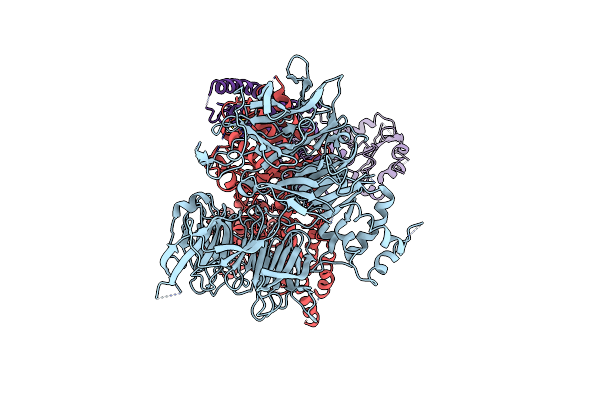 |
Cryo-Em Structure Of Rcmv-E E27 Bound To Human Ddb1 (Deltabpb) And Full-Length Rat Stat2
Organism: Homo sapiens, Murid betaherpesvirus 8, Rattus norvegicus
Method: ELECTRON MICROSCOPY Release Date: 2022-11-09 Classification: VIRUS Ligands: ZN |
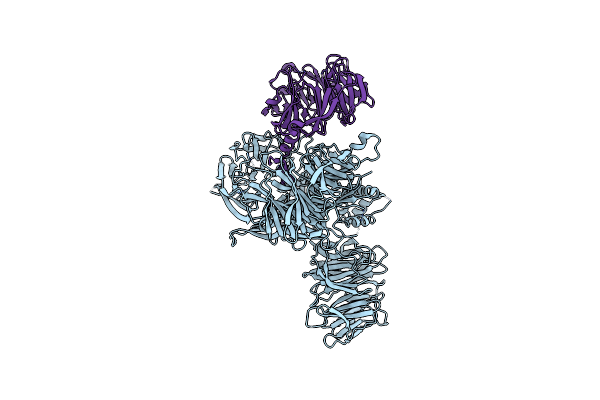 |
Crystal Structure Of Human Ddb1 Bound To Human Dcaf1 (Amino Acid Residues 1046-1396)
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:3.09 Å Release Date: 2021-07-28 Classification: PROTEIN BINDING |
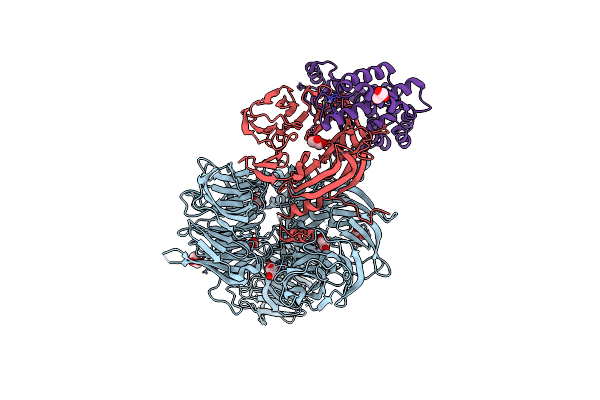 |
Crystal Structure Of Siv Vpr,Fused To T4 Lysozyme, Isolated From Moustached Monkey, Bound To Human Ddb1 And Human Dcaf1 (Amino Acid Residues 1046-1396)
Organism: Homo sapiens, Enterobacteria phage t4, Simian immunodeficiency virus
Method: X-RAY DIFFRACTION Resolution:2.52 Å Release Date: 2021-07-21 Classification: VIRAL PROTEIN Ligands: GOL, ZN |
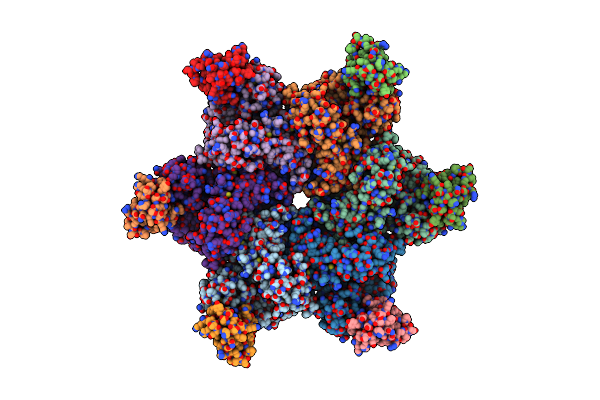 |
Common Mode Of Remodeling Aaa Atpases P97/Cdc48 By Their Disassembly Cofactors Aspl/Pux1
Organism: Homo sapiens, Arabidopsis thaliana
Method: X-RAY DIFFRACTION Resolution:3.73 Å Release Date: 2019-08-28 Classification: GENE REGULATION Ligands: ADP |
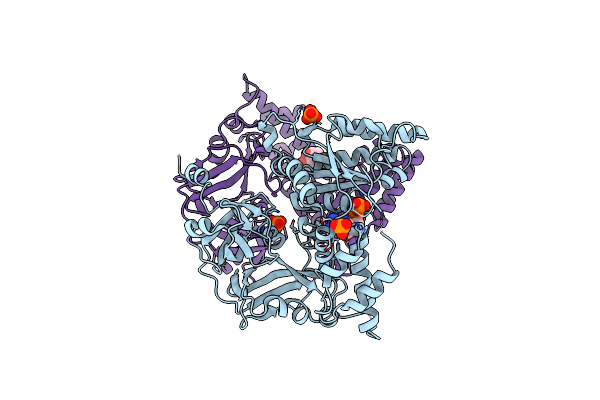 |
Common Mode Of Remodeling Aaa Atpases P97/Cdc48 By Their Disassembly Cofactors Aspl/Pux1
Organism: Arabidopsis thaliana
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2019-08-28 Classification: PLANT PROTEIN Ligands: PO4, ADP |
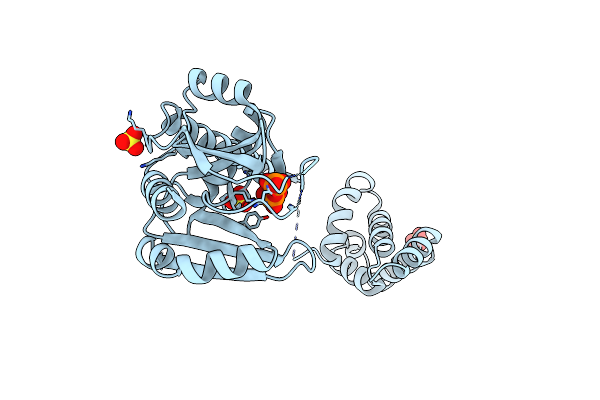 |
Organism: Plasmid rsf1010
Method: X-RAY DIFFRACTION Resolution:1.99 Å Release Date: 2009-04-28 Classification: REPLICATION Ligands: SO4, DPO |
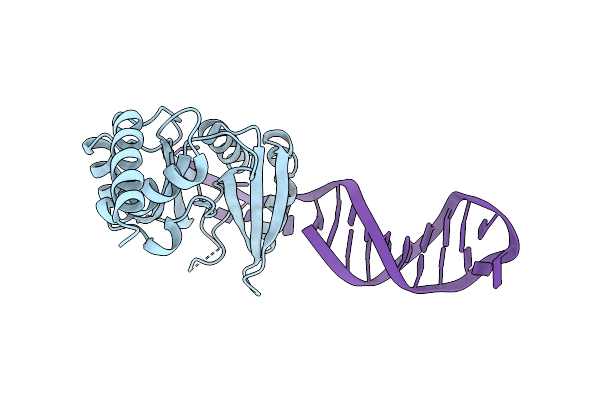 |
Crystal Structure Of The Catalytic Domain Of Primase Repb' In Complex With Initiator Dna
Organism: Plasmid rsf1010
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2009-04-28 Classification: REPLICATION/DNA |

