Search Count: 76
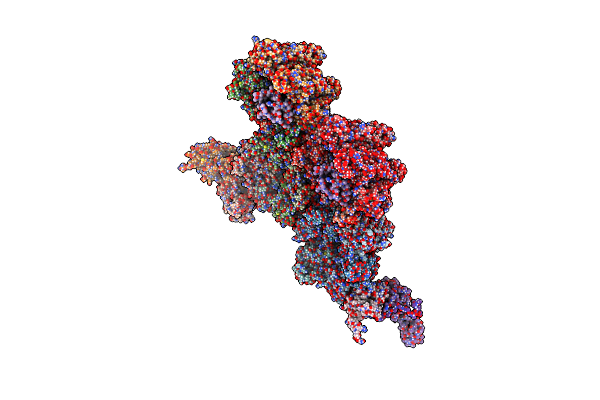 |
Cryo-Em Structure Of The Archaeal Extremophilic Internal Membrane Containing Haloarcula Hispanica Icosahedral Virus 2 (Hhiv-2) At 3.78 Angstroms Resolution.
Organism: Haloarcula hispanica icosahedral virus 2
Method: ELECTRON MICROSCOPY Resolution:3.78 Å Release Date: 2019-04-03 Classification: VIRUS |
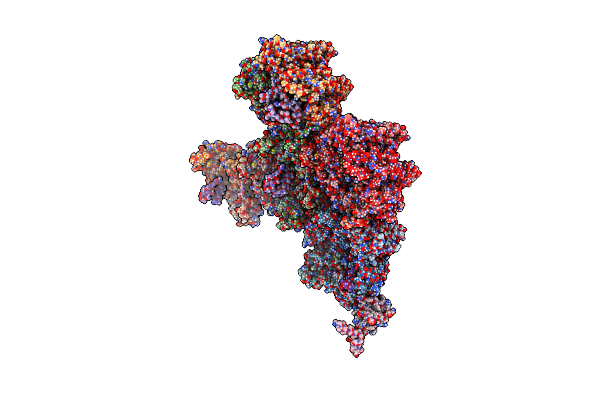 |
Cryo-Em Structure Of Archaeal Extremophilic Internal Membrane-Containing Haloarcula Californiae Icosahedral Virus 1 (Hciv-1) At 3.74 Angstroms Resolution.
Organism: Haloarcula californiae atcc 33799
Method: ELECTRON MICROSCOPY Resolution:3.74 Å Release Date: 2019-03-27 Classification: VIRUS |
 |
Organism: Halorubrum pleomorphic virus 6
Method: ELECTRON MICROSCOPY Release Date: 2019-03-06 Classification: VIRAL PROTEIN |
 |
Organism: Halorubrum pleomorphic virus 2
Method: X-RAY DIFFRACTION Resolution:2.46 Å Release Date: 2019-02-27 Classification: VIRAL PROTEIN Ligands: NAG, CL |
 |
Organism: Halorubrum pleomorphic virus 6
Method: X-RAY DIFFRACTION Resolution:2.69 Å Release Date: 2019-02-27 Classification: VIRAL PROTEIN Ligands: BR |
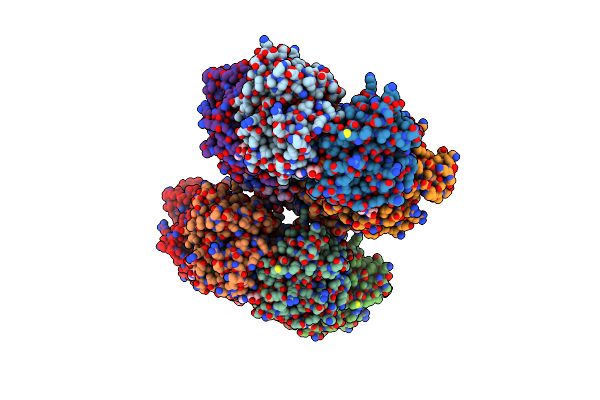 |
Organism: Pseudomonas phage phi6
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2013-08-21 Classification: HYDROLASE Ligands: CA, ADP |
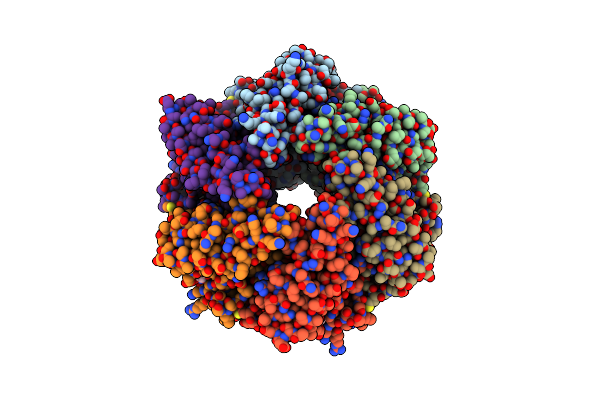 |
Organism: Pseudomonas phage phi13
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2013-08-21 Classification: HYDROLASE Ligands: FLC, GOL |
 |
Organism: Pseudomonas phage phi8
Method: X-RAY DIFFRACTION Resolution:2.79 Å Release Date: 2013-08-21 Classification: HYDROLASE |
 |
Organism: Pseudomonas phage phi12
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2013-08-21 Classification: HYDROLASE Ligands: UTP, DTT |
 |
Organism: Pseudomonas phage phi12
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2013-08-21 Classification: HYDROLASE Ligands: APC |
 |
Organism: Pseudomonas phage phi12
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2013-08-21 Classification: HYDROLASE Ligands: APC |
 |
Organism: Pseudomonas phage phi8
Method: X-RAY DIFFRACTION Resolution:3.10 Å Release Date: 2013-08-21 Classification: HYDROLASE |
 |
Organism: Pseudomonas phage phi8
Method: X-RAY DIFFRACTION Resolution:3.70 Å Release Date: 2013-08-07 Classification: VIRAL PROTEIN |
 |
Organism: Pseudomonas phage phi8
Method: ELECTRON MICROSCOPY Resolution:8.70 Å Release Date: 2013-08-07 Classification: VIRUS |
 |
Organism: Infectious pancreatic necrosis virus
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2013-01-16 Classification: VIRAL PROTEIN Ligands: GOL, K |
 |
The C-Terminal Priming Domain Is Strongly Associated With The Main Body Of Bacteriophage Phi6 Rna-Dependent Rna Polymerase
Organism: Pseudomonas phage phi6
Method: X-RAY DIFFRACTION Resolution:3.30 Å Release Date: 2012-08-01 Classification: TRANSFERASE Ligands: MN |
 |
Non-Catalytic Ions Direct The Rna-Dependent Rna Polymerase Of Bacterial Dsrna Virus Phi6 From De Novo Initiation To Elongation
Organism: Pseudomonas phage phi6, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:3.30 Å Release Date: 2012-07-04 Classification: TRANSFERASE Ligands: MG, ATP |
 |
Non-Catalytic Ions Direct The Rna-Dependent Rna Polymerase Of Bacterial Dsrna Virus Phi6 From De Novo Initiation To Elongation
Organism: Pseudomonas phage phi6, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:3.50 Å Release Date: 2012-07-04 Classification: TRANSFERASE/DNA Ligands: MG, GTP |
 |
Non-Catalytic Ions Direct The Rna-Dependent Rna Polymerase Of Bacterial Dsrna Virus Phi6 From De Novo Initiation To Elongation
Organism: Pseudomonas phage phi6
Method: X-RAY DIFFRACTION Resolution:2.92 Å Release Date: 2012-07-04 Classification: TRANSFERASE/DNA Ligands: MG, GTP, ATP |
 |
Non-Catalytic Ions Direct The Rna-Dependent Rna Polymerase Of Bacterial Dsrna Virus Phi6 From De Novo Initiation To Elongation
Organism: Pseudomonas phage phi6
Method: X-RAY DIFFRACTION Resolution:2.67 Å Release Date: 2012-07-04 Classification: TRANSFERASE Ligands: MN |

