Planned Maintenance: Some services may turn out to be unavailable from 15th January, 2026 to 16th January, 2026. We apologize for the inconvenience!
Planned Maintenance: Some services may turn out to be unavailable from 15th January, 2026 to 16th January, 2026. We apologize for the inconvenience!
 |
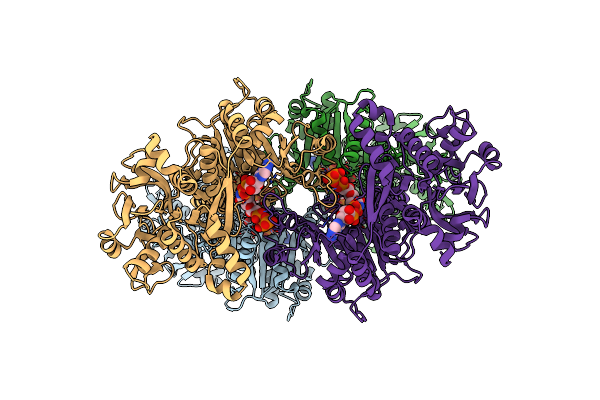
X-Ray Crystal Structure Of The "Closed" Conformation Of Ctp-Inhibited E. Coli Cytidine Triphosphate (Ctp) Synthetase
Organism: Escherichia coli (strain k12)
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2017-04-26 Classification: LYASE Ligands: GLN, SO4, MPD, MG, CTP, MRD |
 |
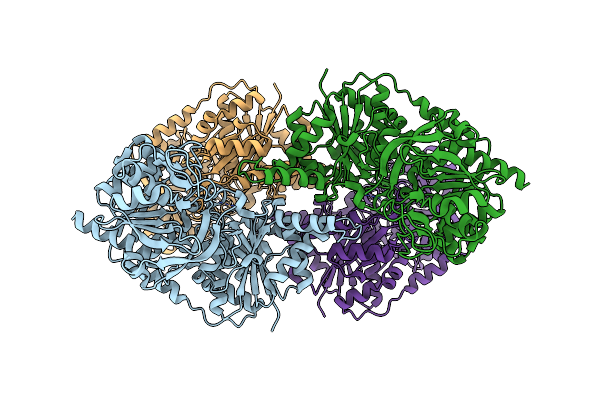
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2017-04-26 Classification: LIGASE, PROTEIN FIBRIL Ligands: ATP, UTP |
 |
Organism: Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2017-04-26 Classification: LIGASE |
 |
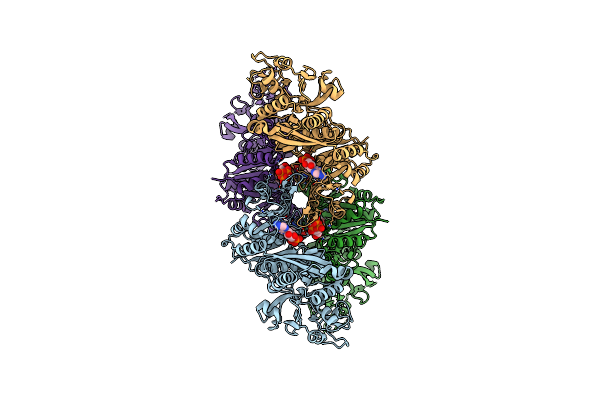
Organism: Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2017-04-26 Classification: PROTEIN FIBRIL, LIGASE Ligands: CTP, ADP |
 |
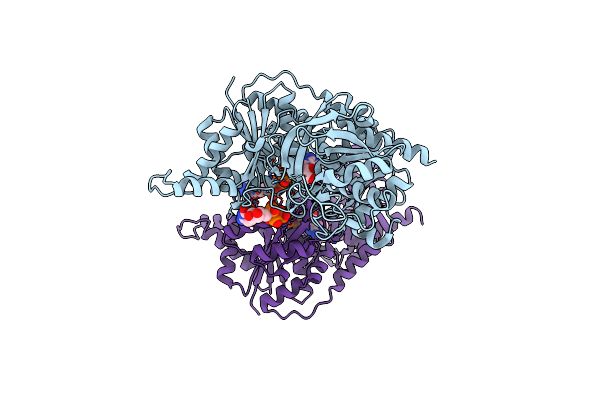
Organism: Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2017-04-26 Classification: PROTEIN FIBRIL, LIGASE Ligands: CTP, ADP |
 |
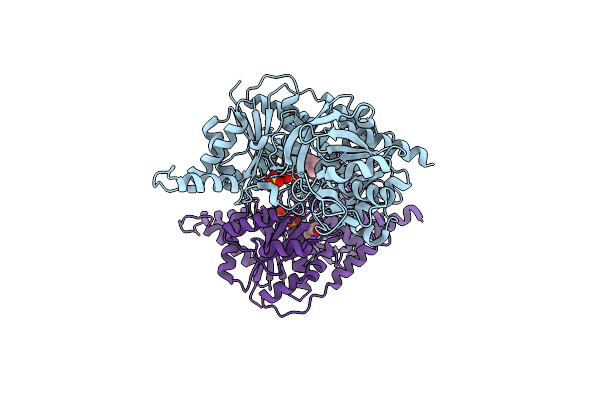
Mechanisms Of Feedback Regulation And Drug Resistance Of Ctp Synthetases: Structure Of The E. Coli Ctps/Ctp Complex At 2.8-Angstrom Resolution.
Organism: Escherichia coli k12
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2005-11-01 Classification: LIGASE Ligands: MG, ADP, CTP |
 |
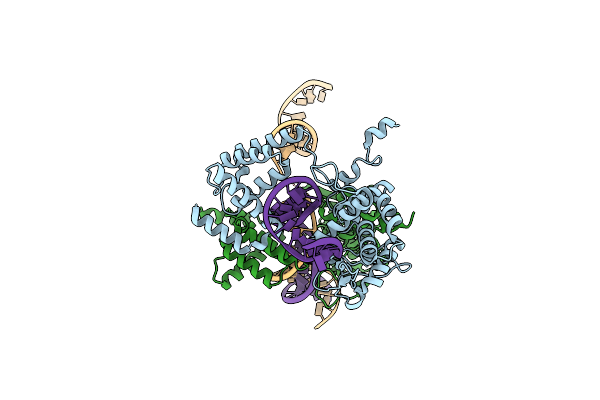
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2004-06-15 Classification: LIGASE Ligands: SO4, MG, IOD, MPD |
 |
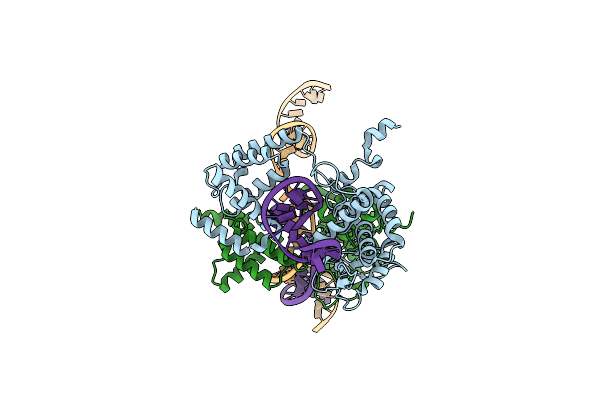
Basis For A Switch In Substrate Specificity: Crystal Structure Of Selected Variant Of Cre Site-Specific Recombinase, Alshg Bound To The Engineered Recognition Site Loxm7
Organism: Escherichia phage p1, Escherichia virus p1
Method: X-RAY DIFFRACTION Resolution:2.35 Å Release Date: 2004-02-17 Classification: RECOMBINATION/DNA |
 |
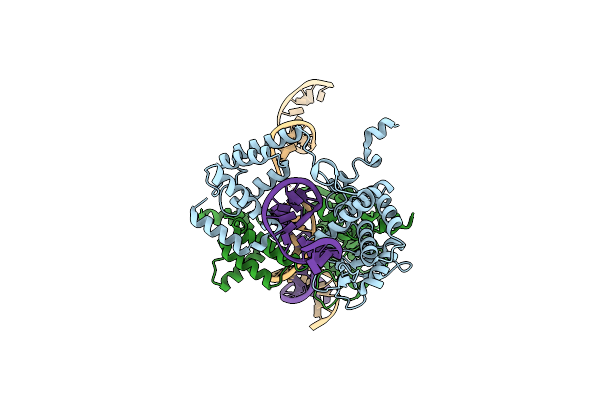
Basis For A Switch In Substrate Specificity: Crystal Structure Of Selected Variant Of Cre Site-Specific Recombinase, Lnsgg Bound To The Engineered Recognition Site Loxm7
Organism: Escherichia phage p1, Escherichia virus p1
Method: X-RAY DIFFRACTION Resolution:2.75 Å Release Date: 2004-02-17 Classification: RECOMBINATION/DNA |
 |
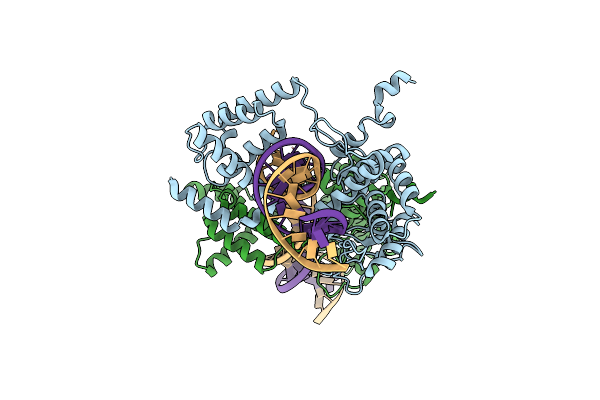
Basis For A Switch In Substrate Specificity: Crystal Structure Of Selected Variant Of Cre Site-Specific Recombinase, Lnsgg Bound To The Loxp (Wildtype) Recognition Site
Organism: Enterobacteria phage p1
Method: X-RAY DIFFRACTION Resolution:2.65 Å Release Date: 2004-02-17 Classification: RECOMBINATION/DNA |
 |
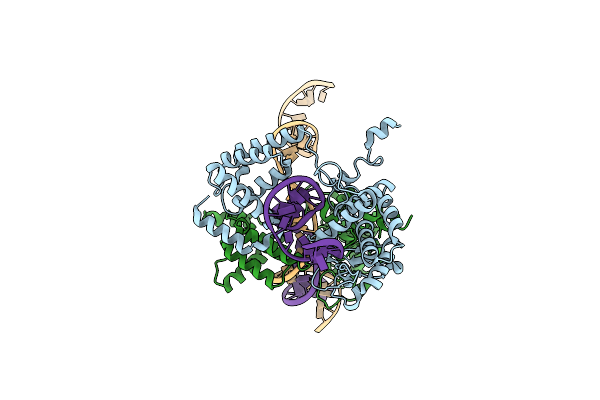
Crystal Structure Of Cre Site-Specific Recombinase Complexed With A Mutant Dna Substrate, Loxp-A8/T27
Organism: Enterobacteria phage p1
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2003-07-08 Classification: HYDROLASE, LIGASE/DNA |
 |
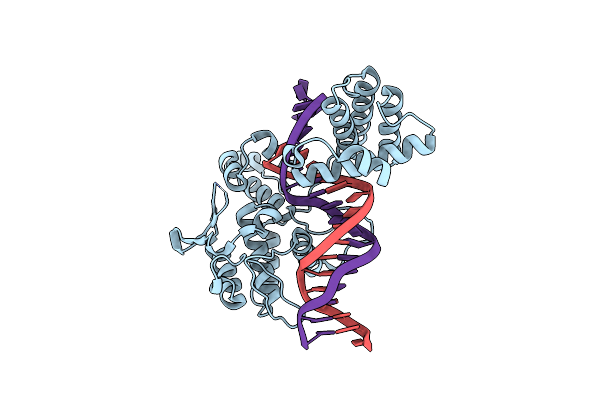
Organism: Enterobacteria phage p1
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2002-06-07 Classification: HYDROLASE, LIGASE/DNA |
 |
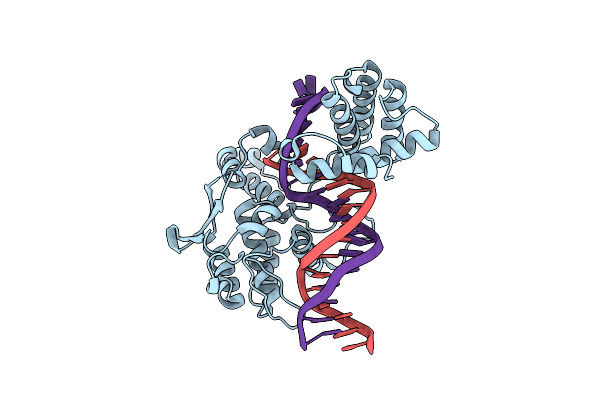
Organism: Enterobacteria phage p1
Method: X-RAY DIFFRACTION Resolution:2.55 Å Release Date: 2001-10-19 Classification: HYDROLASE, LIGASE/DNA |
 |
Organism: Enterobacteria phage p1
Method: X-RAY DIFFRACTION Resolution:2.05 Å Release Date: 2001-10-12 Classification: HYDROLASE, LIGASE/DNA |
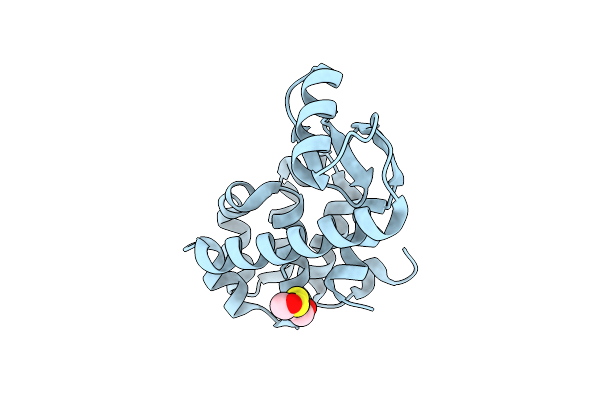 |
Organism: Enterobacteria phage t4
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2001-05-23 Classification: HYDROLASE Ligands: CL, HED |
 |
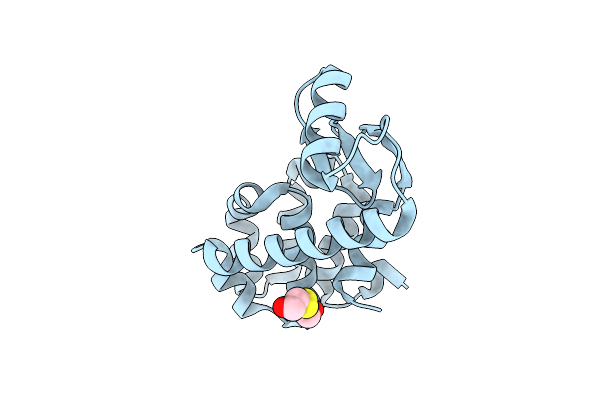
Organism: Enterobacteria phage t4
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2001-05-23 Classification: HYDROLASE Ligands: CL, HED |
 |
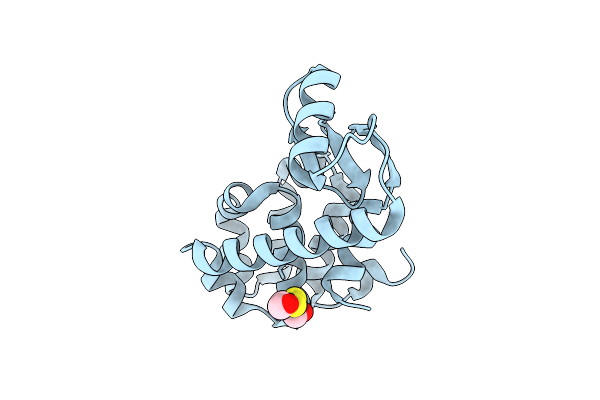
Organism: Enterobacteria phage t4
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2001-05-23 Classification: HYDROLASE Ligands: CL, HED |
 |
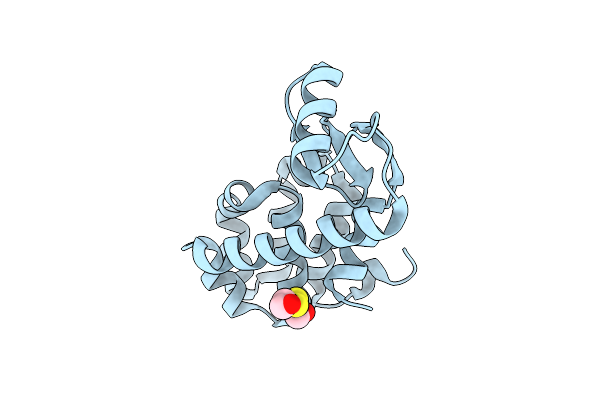
Organism: Enterobacteria phage t4
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2001-05-23 Classification: HYDROLASE Ligands: CL, HED |
 |
Organism: Enterobacteria phage t4
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2001-05-23 Classification: HYDROLASE Ligands: CL, HED |
 |
Organism: Enterobacteria phage t4
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2001-05-23 Classification: HYDROLASE Ligands: CL, HED |