Search Count: 66
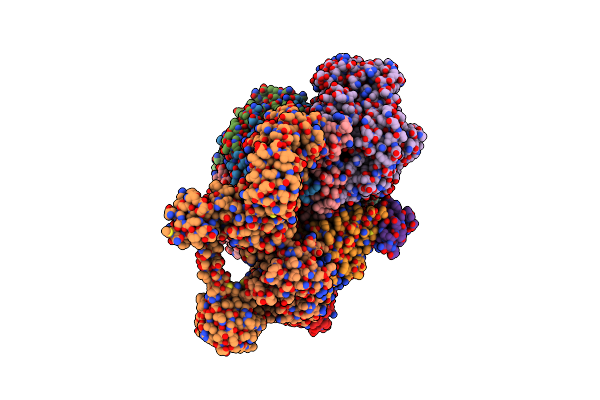 |
Organism: Methanococcus maripaludis
Method: ELECTRON MICROSCOPY Release Date: 2025-02-26 Classification: OXIDOREDUCTASE Ligands: SHT, TP7, COM, F43, S5Q, ZN, ATP, MG |
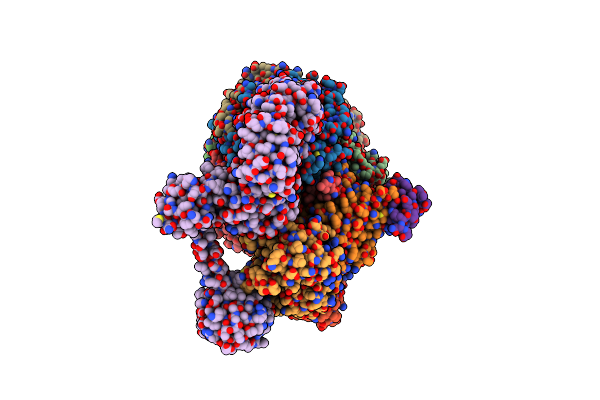 |
Organism: Methanococcus maripaludis
Method: ELECTRON MICROSCOPY Release Date: 2025-02-26 Classification: OXIDOREDUCTASE Ligands: F43, SHT, COM, TP7, S5Q |
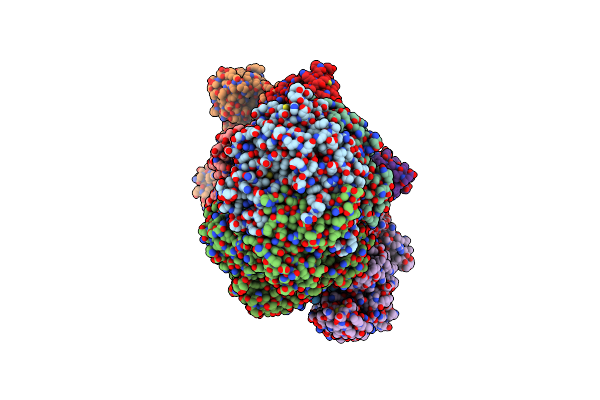 |
Methyl-Coenzyme M Reductase Activation Complex Binding To The A2 Component After Incubation With Atp
Organism: Methanococcus maripaludis
Method: ELECTRON MICROSCOPY Release Date: 2025-02-26 Classification: OXIDOREDUCTASE Ligands: F43, COM, TP7, SHT, S5Q, ZN, ATP, MG |
 |
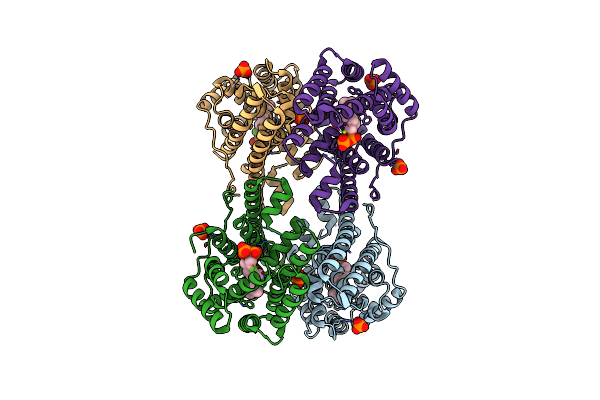
Crystal Structure Of Caryolan-1-Ol Synthase From S. Griseus With Peg Molecule In The Active Site
Organism: Streptomyces griseus
Method: X-RAY DIFFRACTION Resolution:2.33 Å Release Date: 2024-11-06 Classification: METAL BINDING PROTEIN Ligands: PG4, CA, 1PE |
 |
Crystal Structure Of Caryolan-1-Ol Synthase Complexed With 2-Fluorofarnesyl Diphosphate
Organism: Streptomyces griseus
Method: X-RAY DIFFRACTION Resolution:2.65 Å Release Date: 2024-11-06 Classification: METAL BINDING PROTEIN Ligands: FPF, PO4, MG, DPO |
 |
Crystal Structure Of Pentalenene Synthase Variant F76A With Peg Molecule In The Active Site
Organism: Streptomyces exfoliatus
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2024-11-06 Classification: METAL BINDING PROTEIN Ligands: PG4, SO4 |
 |
Crystal Structure Of Pentalenene Synthase Variant F76A Complexed With 2-Fluorofarnesyl Diphosphate
Organism: Streptomyces exfoliatus
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2024-11-06 Classification: METAL BINDING PROTEIN Ligands: FPF, MG |
 |
Crystal Structure Of Pentalenene Synthase Variant F76A Complexed With 12,13-Difluorofarnesyl Diphosphate
Organism: Streptomyces exfoliatus
Method: X-RAY DIFFRACTION Resolution:2.65 Å Release Date: 2024-11-06 Classification: METAL BINDING PROTEIN Ligands: FDF, MG |
 |
Organism: Bombyx mori
Method: ELECTRON MICROSCOPY Release Date: 2024-01-10 Classification: MEMBRANE PROTEIN Ligands: PSC, PC7, 9Z9 |
 |
Cryoem Structure Of Insect Gustatory Receptor Bmgr9 In The Presence Of Fructose
Organism: Bombyx mori
Method: ELECTRON MICROSCOPY Release Date: 2024-01-10 Classification: MEMBRANE PROTEIN Ligands: PSC |
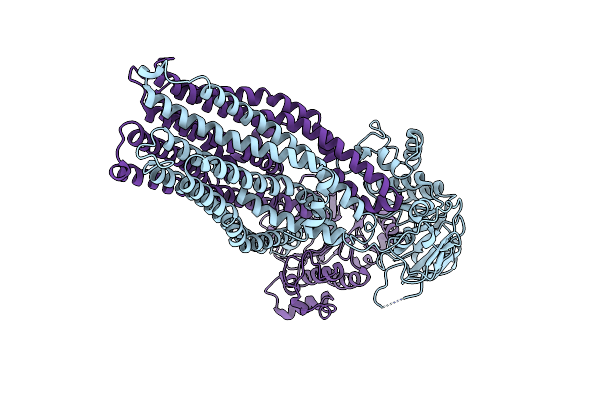 |
Organism: Escherichia coli k-12
Method: ELECTRON MICROSCOPY Release Date: 2023-04-19 Classification: MEMBRANE PROTEIN |
 |
If(Apo/Asym) Conformation Of Cyddc In Adp+Pi(Cydc)/Atp(Cydd) Bound State (Dataset-2)
Organism: Escherichia coli k-12
Method: ELECTRON MICROSCOPY Release Date: 2023-04-19 Classification: MEMBRANE PROTEIN Ligands: MG, ADP, PO3, ATP |
 |
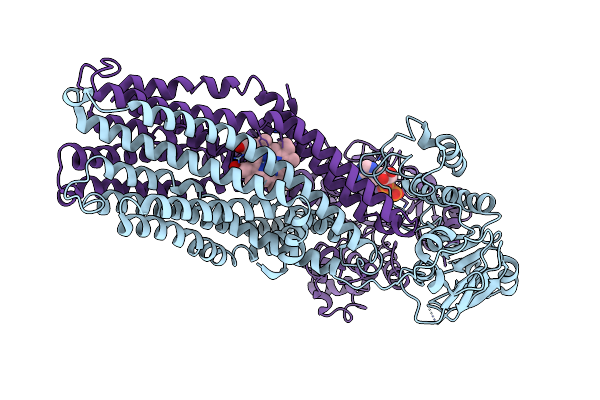
If(Heme/Bound) Conformation Of Cyddc In Adp+Pi(Cydc)/Atp(Cydd) Bound State (Dataset-2)
Organism: Escherichia coli k-12
Method: ELECTRON MICROSCOPY Release Date: 2023-04-19 Classification: MEMBRANE PROTEIN Ligands: HEB, MG, ADP, PO3, ATP |
 |
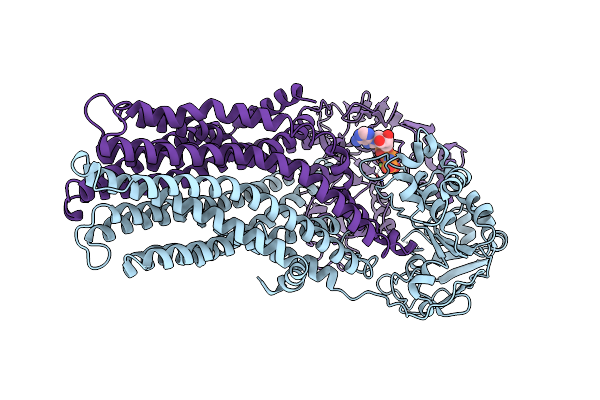
If(Heme/Confined) Conformation Of Cyddc In Adp(Cydd) Bound State (Dataset-3)
Organism: Escherichia coli k-12
Method: ELECTRON MICROSCOPY Release Date: 2023-04-19 Classification: MEMBRANE PROTEIN Ligands: HEB, MG, ADP |
 |
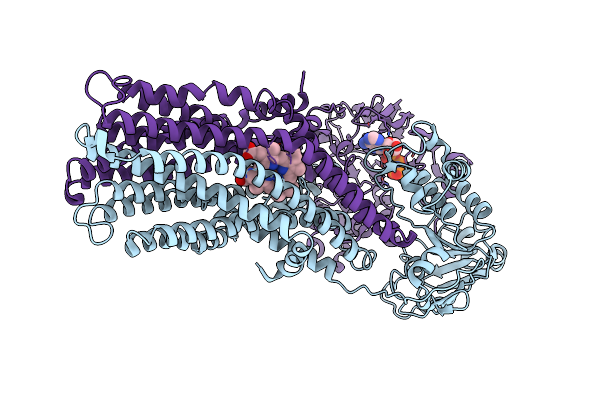
If(Apo/As Isolated) Conformation Of Cyddc In Amp-Pnp(Cydd) Bound State (Dataset-4)
Organism: Escherichia coli k-12
Method: ELECTRON MICROSCOPY Release Date: 2023-04-19 Classification: MEMBRANE PROTEIN Ligands: MG, ANP |
 |
If(Heme/Confined) Conformation Of Cyddc In Amp-Pnp(Cydd) Bound State (Dataset-4)
Organism: Escherichia coli k-12
Method: ELECTRON MICROSCOPY Release Date: 2023-04-19 Classification: MEMBRANE PROTEIN Ligands: ISW, MG, ANP |
 |
Organism: Escherichia coli k-12
Method: ELECTRON MICROSCOPY Release Date: 2023-04-19 Classification: MEMBRANE PROTEIN Ligands: HEB |
 |
If(Apo/Asym) Conformation Of Cyddc In Amp-Pnp(Cydc)/Amp-Pnp(Cydd) Bound State (Dataset-8)
Organism: Escherichia coli k-12
Method: ELECTRON MICROSCOPY Release Date: 2023-04-19 Classification: MEMBRANE PROTEIN Ligands: MG, ANP |
 |
If(Heme/Coordinated) Conformation Of Cyddc In Amp-Pnp(Cydc)/Amp-Pnp(Cydd) Bound State (Dataset-8)
Organism: Escherichia coli k-12
Method: ELECTRON MICROSCOPY Release Date: 2023-04-19 Classification: MEMBRANE PROTEIN Ligands: HEB, MG, ANP |
 |
If(Apo/As Isolated) Conformation Of Cyddc Mutant (H85A.C) In Amp-Pnp(Cydd) Bound State (Dataset-16)
Organism: Escherichia coli k-12
Method: ELECTRON MICROSCOPY Release Date: 2023-04-19 Classification: MEMBRANE PROTEIN Ligands: MG, ANP |

