Search Count: 36
 |
Organism: Gloeobacter violaceus
Method: ELECTRON MICROSCOPY Release Date: 2024-04-17 Classification: MEMBRANE PROTEIN Ligands: PEE |
 |
Cryo-Em Structure Of Nanodisc (Pe:Ps:Pc) Reconstituted Glic At Ph 4 In Closed State
Organism: Gloeobacter violaceus
Method: ELECTRON MICROSCOPY Release Date: 2024-04-17 Classification: MEMBRANE PROTEIN Ligands: CL, PEE |
 |
Organism: Gloeobacter violaceus
Method: ELECTRON MICROSCOPY Release Date: 2024-04-17 Classification: MEMBRANE PROTEIN Ligands: CL, PEE |
 |
Cryo-Em Structure Of Nanodisc (Pe:Ps:Pc) Reconstituted Glic At Ph 4 In Intermediate State
Organism: Gloeobacter violaceus
Method: ELECTRON MICROSCOPY Release Date: 2024-04-17 Classification: MEMBRANE PROTEIN Ligands: CL, PEE |
 |
Cryo-Em Structure Of Nanodisc (Pe:Ps:Pc) Reconstituted Glic At Ph 4 In Open State
Organism: Gloeobacter violaceus
Method: ELECTRON MICROSCOPY Release Date: 2024-04-17 Classification: MEMBRANE PROTEIN Ligands: CL, PEE |
 |
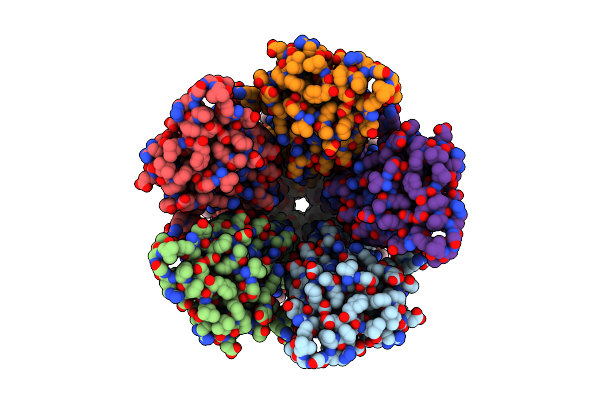
Organism: Gloeobacter violaceus
Method: ELECTRON MICROSCOPY Release Date: 2024-04-10 Classification: MEMBRANE PROTEIN Ligands: PLC |
 |
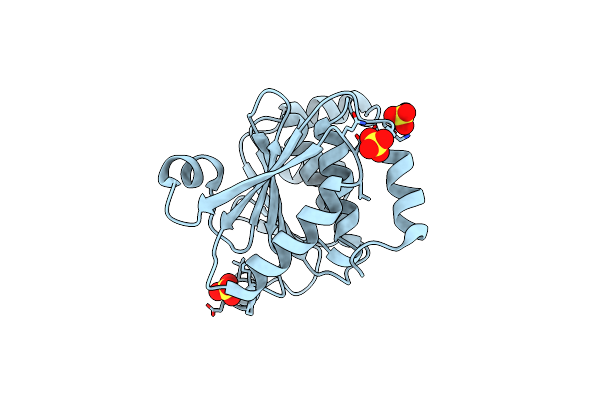
Organism: Gloeobacter violaceus
Method: ELECTRON MICROSCOPY Release Date: 2024-04-10 Classification: MEMBRANE PROTEIN Ligands: PEE |
 |
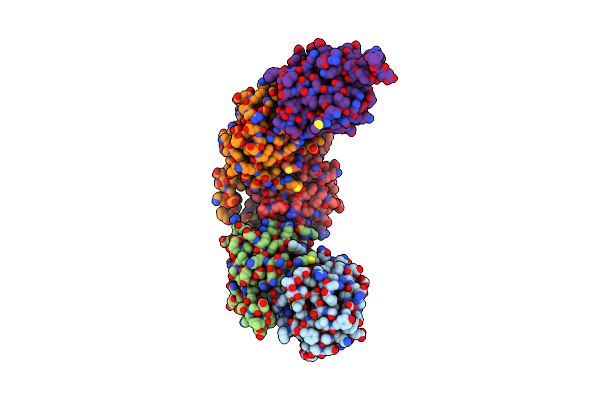
Structure Of Enterococcus Faecalis (V583) Alkylhydroperoxide Reductase Subunit F (Ahpf) C503A Mutant
Organism: Enterococcus faecalis v583
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2019-05-22 Classification: OXIDOREDUCTASE Ligands: SO4 |
 |
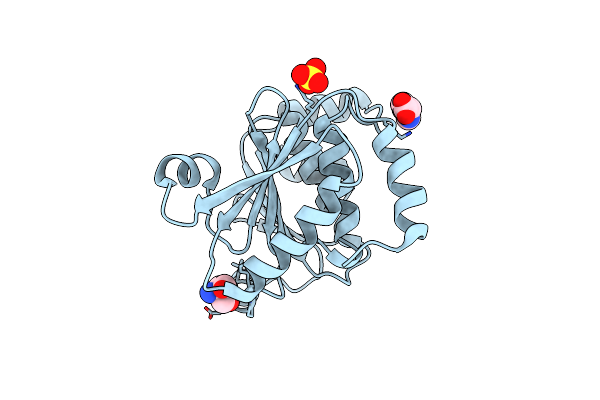
Organism: Enterococcus faecalis (strain atcc 700802 / v583)
Method: X-RAY DIFFRACTION Resolution:2.87 Å Release Date: 2017-12-27 Classification: OXIDOREDUCTASE |
 |
Crystal Structure Of The Ntd_N/C Domain Of Alkylhydroperoxide Reductase Ahpf From Enterococcus Faecalis (V583)
Organism: Enterococcus faecalis
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2017-11-22 Classification: OXIDOREDUCTASE Ligands: SO4, TRS, PRO |
 |
Crystallographic Structure Of The Enzymatically Active N-Terminal Domain Of The Rel Protein From Mycobacterium Tuberculosis
Organism: Mycobacterium tuberculosis (strain atcc 25618 / h37rv)
Method: X-RAY DIFFRACTION Resolution:3.70 Å Release Date: 2017-07-19 Classification: HYDROLASE, TRANSFERASE Ligands: MG |
 |
Asymmetry In The Active Site Of Mycobacterium Tuberculosis Ahpe Upon Exposure To Mycothiol
Organism: Mycobacterium tuberculosis h37rv
Method: X-RAY DIFFRACTION Resolution:2.43 Å Release Date: 2016-08-03 Classification: OXIDOREDUCTASE Ligands: GOL, ACT |
 |
Organism: Saccharomyces cerevisiae s288c
Method: X-RAY DIFFRACTION Resolution:3.18 Å Release Date: 2014-12-10 Classification: HYDROLASE Ligands: GOL |
 |
Crystal Structure Of The Alkylhydroperoxide Reductase Ahpf From Escherichia Coli
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2014-11-05 Classification: OXIDOREDUCTASE Ligands: FAD, SO4, PGE, GOL, CD, PG4, TRS |
 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:3.33 Å Release Date: 2014-11-05 Classification: OXIDOREDUCTASE |
 |
Crystal Structure Of Alkylhydroperoxide Reductase Subunit F From E. Coli At 2.65 Ang Resolution
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.65 Å Release Date: 2014-11-05 Classification: OXIDOREDUCTASE Ligands: SO4, FAD, CD |
 |
Organism: Triticum aestivum
Method: X-RAY DIFFRACTION Resolution:6.00 Å Release Date: 2014-03-19 Classification: HYDROLASE |
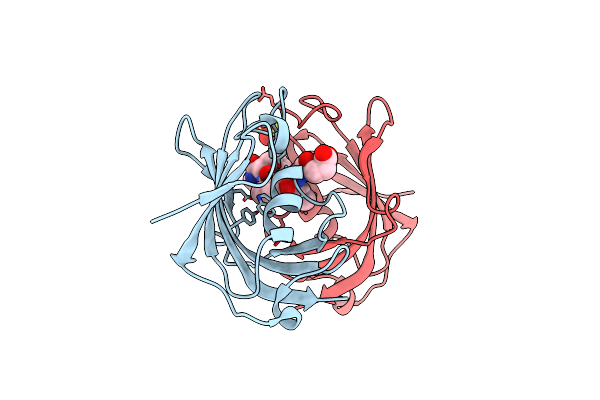 |
Crystal Structure Of The Fk506 Binding Domain Of Plasmodium Vivax Fkbp35 In Complex With A Tetrapeptide Substrate
Organism: Plasmodium vivax
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2013-03-20 Classification: ISOMERASE/SUBSTRATE Ligands: SO4 |
 |
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:2.33 Å Release Date: 2013-03-20 Classification: HYDROLASE Ligands: TRS, GOL |
 |
Organism: Pyrococcus horikoshii
Method: X-RAY DIFFRACTION Resolution:3.65 Å Release Date: 2012-05-02 Classification: HYDROLASE |

