Search Count: 14
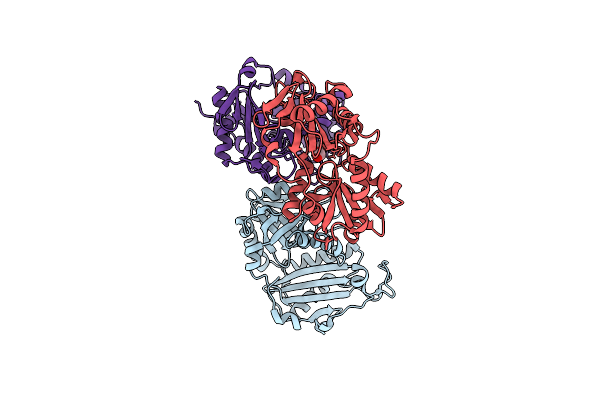 |
Crystal Structure Of Aminotransferase-Like Protein From Variovorax Paradoxus Mutant N174K
Organism: Variovorax paradoxus b4
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2025-01-29 Classification: TRANSFERASE Ligands: PEG |
 |
Crystal Structure Of Monomeric Plp-Dependent Transaminase From Desulfobacula Toluolica In F 41 3 2 Space Group
Organism: Desulfobacula toluolica
Method: X-RAY DIFFRACTION Resolution:2.58 Å Release Date: 2024-09-18 Classification: TRANSFERASE |
 |
Crystal Structure Of Monomeric Plp-Dependent Transaminase From Desulfobacula Toluolica In P 21 21 21 Space Group
Organism: Desulfobacula toluolica
Method: X-RAY DIFFRACTION Resolution:1.79 Å Release Date: 2024-09-18 Classification: TRANSFERASE Ligands: PEG, GOL |
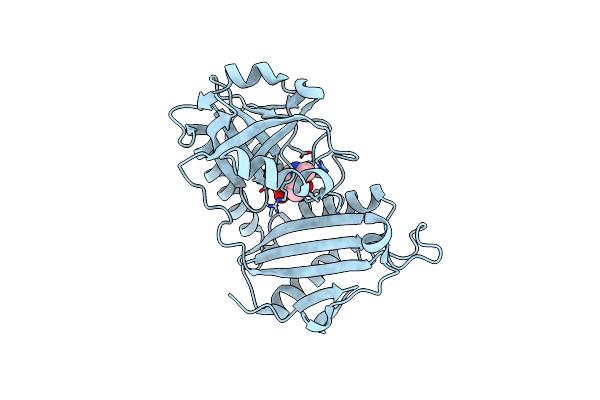 |
Crystal Structure Of D-Amino Acid Transaminase From Haliscomenobacter Hydrossis In The Holo Form Obtained At Ph 7.0
Organism: Haliscomenobacter hydrossis dsm 1100
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2024-04-17 Classification: TRANSFERASE Ligands: PLP |
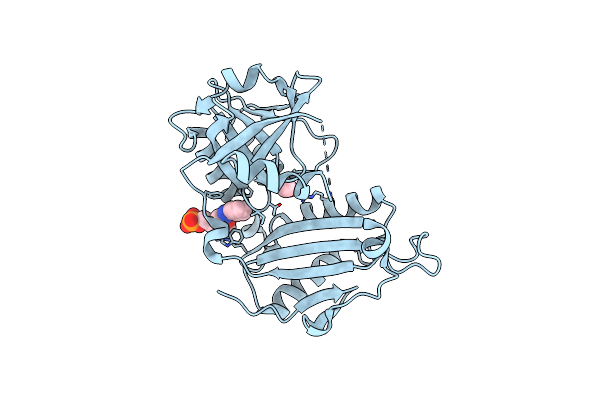 |
Crystal Structure Of D-Amino Acid Transaminase From Haliscomenobacter Hydrossis (Apo Form) After 15 Sec Of Soaking With Phenylhydrazine
Organism: Haliscomenobacter hydrossis dsm 1100
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2024-04-17 Classification: TRANSFERASE Ligands: ACT, ZXN |
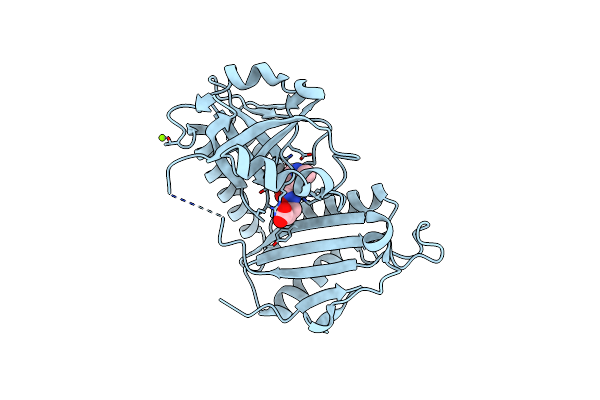 |
Crystal Structure Of D-Amino Acid Transaminase From Haliscomenobacter Hydrossis Complexed With 3-Aminooxypropionic Acid
Organism: Haliscomenobacter hydrossis dsm 1100
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2024-04-17 Classification: TRANSFERASE Ligands: OCF, MG |
 |
Crystal Structure Of D-Amino Acid Transaminase From Haliscomenobacter Hydrossis Point Mutant R90I (Holo Form)
Organism: Haliscomenobacter hydrossis dsm 1100
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2023-12-27 Classification: TRANSFERASE Ligands: PLP |
 |
Crystal Structure Of D-Amino Acid Transaminase From Haliscomenobacter Hydrossis Point Mutant R90I Complexed With Phenylhydrazine
Organism: Haliscomenobacter hydrossis dsm 1100
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2023-12-27 Classification: TRANSFERASE Ligands: ZXN, GOL |
 |
Crystal Structure Of D-Amino Acid Aminotrensferase From Haliscomenobacter Hydrossis Complexed With D-Cycloserine
Organism: Haliscomenobacter hydrossis
Method: X-RAY DIFFRACTION Resolution:1.41 Å Release Date: 2022-08-31 Classification: TRANSFERASE Ligands: GOL, LCS |
 |
Crystal Structure Of D-Aminoacid Transaminase From Haliscomenobacter Hydrossis In Its Intermediate Form
Organism: Haliscomenobacter hydrossis dsm 1100
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2022-08-03 Classification: TRANSFERASE Ligands: SO4, MG |
 |
Crystal Structure Of D-Amino Acid Transaminase From Haliscomenobacter Hydrossis (Holo Form).
Organism: Haliscomenobacter hydrossis (strain atcc 27775 / dsm 1100 / lmg 10767 / o)
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2021-07-28 Classification: TRANSFERASE Ligands: PLP, PO4, ACT |
 |
Crystal Structure Of Branched-Chain Amino Acid Aminotransferase From Thermobaculum Terrenum (M3 Mutant).
Organism: Thermobaculum terrenum (strain atcc baa-798 / ynp1)
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2021-07-07 Classification: TRANSFERASE Ligands: PLP, CL, GOL, NA |
 |
Crystal Structure Of Branched-Chain Amino Acid Aminotransferase From Thermobaculum Terrenum (M4 Mutant)
Organism: Thermobaculum terrenum (strain atcc baa-798 / ynp1)
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2021-07-07 Classification: TRANSFERASE Ligands: PLP, NA |
 |
Crystal Structure Of Adenosylmethionine-8-Amino-7-Oxononanoate Aminotransferase From Methanocaldococcus Jannaschii Dsm 2661
Organism: Methanocaldococcus jannaschii (strain atcc 43067 / dsm 2661 / jal-1 / jcm 10045 / nbrc 100440)
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2020-07-01 Classification: TRANSFERASE Ligands: MG |

