Search Count: 75
 |
Organism: Escherichia phage ms2
Method: ELECTRON MICROSCOPY Release Date: 2025-08-06 Classification: VIRUS LIKE PARTICLE |
 |
Organism: Escherichia phage ms2
Method: ELECTRON MICROSCOPY Release Date: 2025-08-06 Classification: VIRUS LIKE PARTICLE |
 |
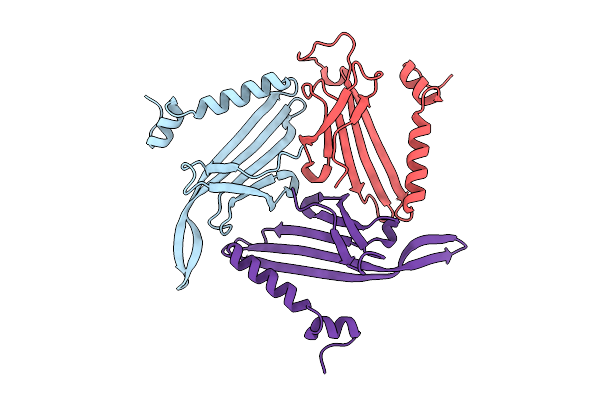
Crystal Structure Of The Tetramerization Domain (29-147) From Human Voltage-Gated Potassium Channel Kv2.1 In P 41 21 2 Space Group
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2022-06-08 Classification: SIGNALING PROTEIN Ligands: ZN |
 |
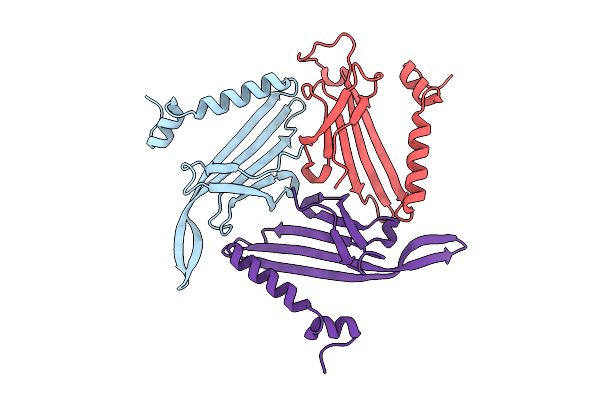
Crystal Structure Of The Tetramerization Domain (29-147) From Human Voltage-Gated Potassium Channel Kv2.1 In C 2 2 21 Space Group
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2022-06-08 Classification: SIGNALING PROTEIN Ligands: PEG, ZN |
 |
Structure Of Human Ck2 Alpha Kinase (Catalytic Subunit) With The Inhibitor 108600.
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2021-08-11 Classification: TRANSFERASE/TRANSFERASE INHIBITOR Ligands: SO4, GOL, ON6 |
 |
The Crystal Structure Of Sars-Cov-2 Main Protease In Complex With Demethylated Analog Of Masitinib
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Resolution:1.58 Å Release Date: 2020-12-30 Classification: HYDROLASE Ligands: XNJ, DMS, GOL |
 |
The Crystal Structure Of Sars-Cov-2 Main Protease In Complex With Masitinib
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2020-09-09 Classification: HYDROLASE Ligands: G65, DMS, GOL |
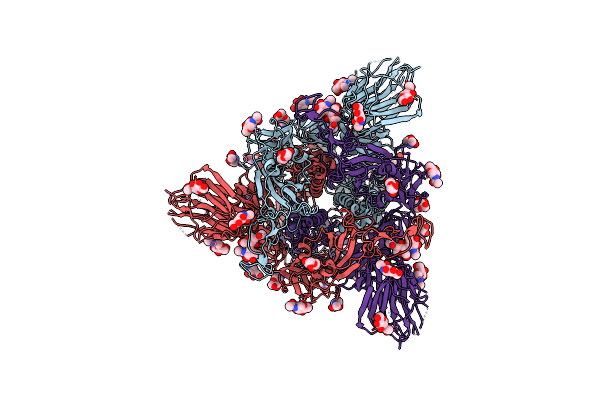 |
Structure Of Disulphide-Stabilized Sars-Cov-2 Spike Protein Trimer (X2 Disulphide-Bond Mutant, G413C, V987C, Single Arg S1/S2 Cleavage Site)
Organism: Severe acute respiratory syndrome coronavirus 2
Method: ELECTRON MICROSCOPY Release Date: 2020-07-22 Classification: VIRAL PROTEIN Ligands: NAG |
 |
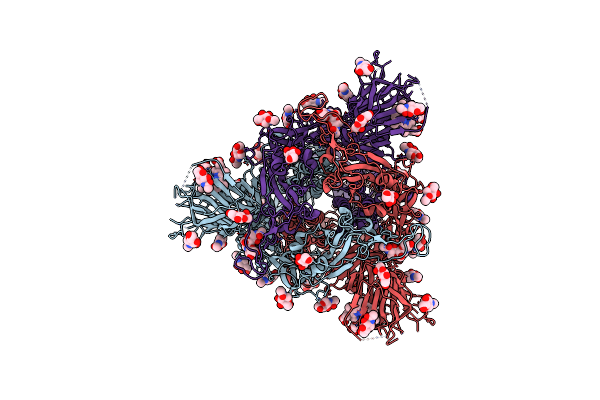
Structure Of Disulphide-Stabilized Sars-Cov-2 Spike Protein Trimer (X1 Disulphide-Bond Mutant, S383C, D985C, K986P, V987P, Single Arg S1/S2 Cleavage Site) In Closed State
Organism: Severe acute respiratory syndrome coronavirus 2
Method: ELECTRON MICROSCOPY Release Date: 2020-07-22 Classification: VIRAL PROTEIN Ligands: NAG |
 |
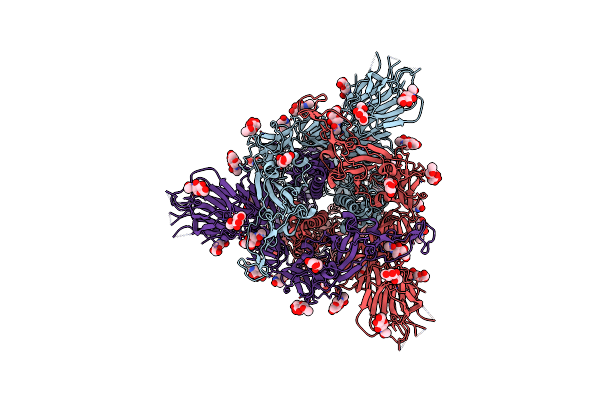
Structure Of Disulphide-Stabilized Sars-Cov-2 Spike Protein Trimer (X1 Disulphide-Bond Mutant, S383C, D985C, K986P, V987P, Single Arg S1/S2 Cleavage Site) In Locked State
Organism: Severe acute respiratory syndrome coronavirus 2
Method: ELECTRON MICROSCOPY Release Date: 2020-07-22 Classification: VIRAL PROTEIN Ligands: NAG, BLA |
 |
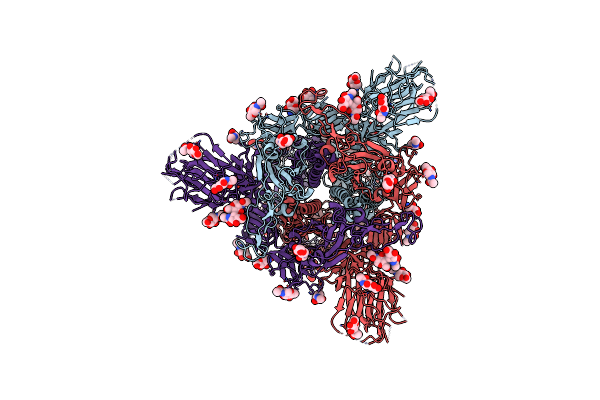
Structure Of Sars-Cov-2 Spike Protein Trimer (Single Arg S1/S2 Cleavage Site) In Closed State
Organism: Severe acute respiratory syndrome coronavirus 2
Method: ELECTRON MICROSCOPY Release Date: 2020-07-22 Classification: VIRAL PROTEIN Ligands: NAG |
 |
Structure Of Sars-Cov-2 Spike Protein Trimer (K986P, V987P, Single Arg S1/S2 Cleavage Site) In Closed State
Organism: Severe acute respiratory syndrome coronavirus 2
Method: ELECTRON MICROSCOPY Release Date: 2020-07-22 Classification: VIRAL PROTEIN Ligands: NAG |
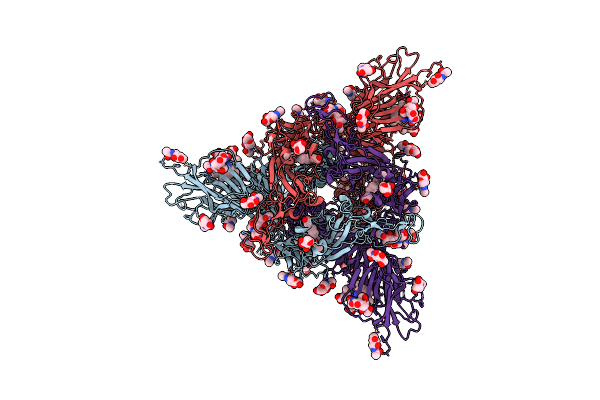 |
Structure Of Sars-Cov-2 Spike Protein Trimer (K986P, V987P, Single Arg S1/S2 Cleavage Site) In Locked State
Organism: Severe acute respiratory syndrome coronavirus 2
Method: ELECTRON MICROSCOPY Release Date: 2020-07-22 Classification: VIRAL PROTEIN Ligands: BLA, NAG, EIC |
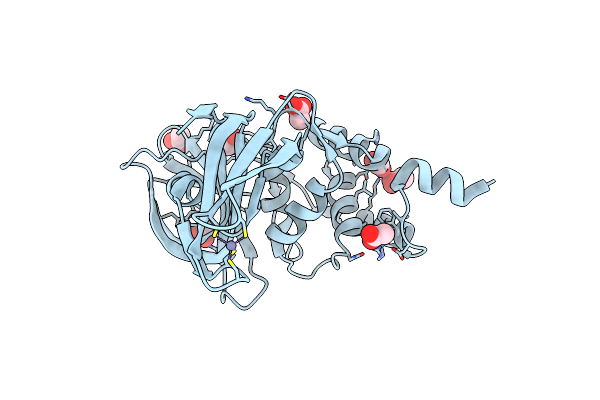 |
Structure Of A Core Papain-Like Protease Of Mers Coronavirus With Utility For Structure-Based Drug Design
Organism: Middle east respiratory syndrome coronavirus
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2017-01-25 Classification: HYDROLASE Ligands: ZN, ACT |
 |
Organism: Homo sapiens
Method: SOLUTION NMR Release Date: 2016-10-05 Classification: PROTEIN BINDING |
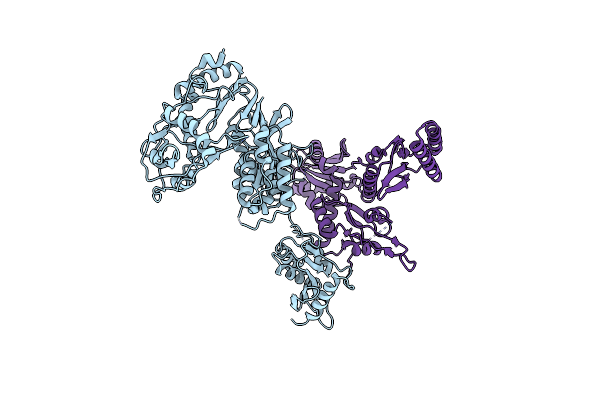 |
Crystal Structure Of Mouse Erf1 In Complex With Reverse Transcriptase (Rt) Of Moloney Murine Leukemia Virus
Organism: Moloney murine leukemia virus (isolate shinnick), Mus musculus
Method: X-RAY DIFFRACTION Resolution:4.00 Å Release Date: 2016-07-06 Classification: TRANSFERASE, HYDROLASE/TRANSLATION |
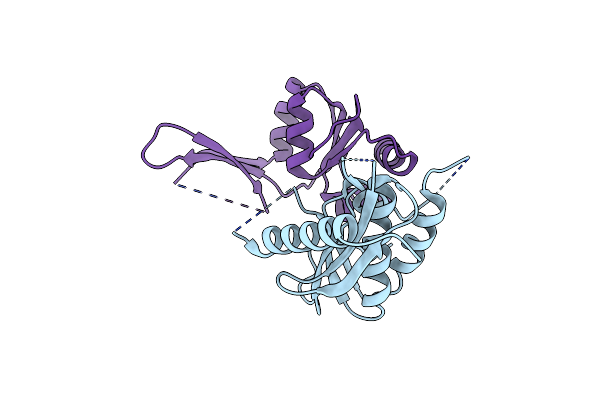 |
Crystal Structure Of C-Terminal Domain Of Mouse Erf1 In Complex With Rnase H Domain Of Rt Of Moloney Murine Leukemia Virus
Organism: Moloney murine leukemia virus (isolate shinnick), Mus musculus
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2016-07-06 Classification: HYDROLASE/TRANSLATION |
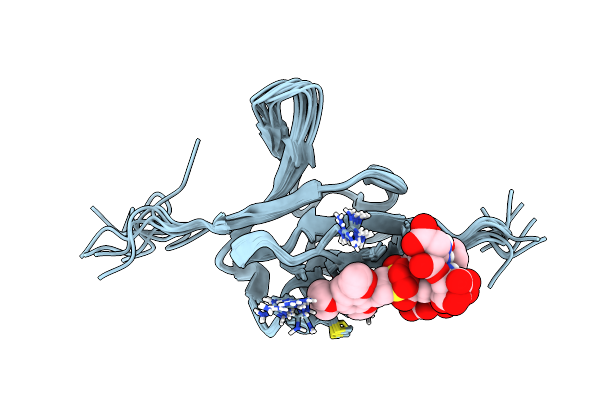 |
Solution Structure Of Ras Binding Domain (Rbd) Of B-Raf Complexed With Rigosertib (Complex I)
Organism: Homo sapiens
Method: SOLUTION NMR Release Date: 2016-05-04 Classification: PROTEIN BINDING Ligands: 6FS |
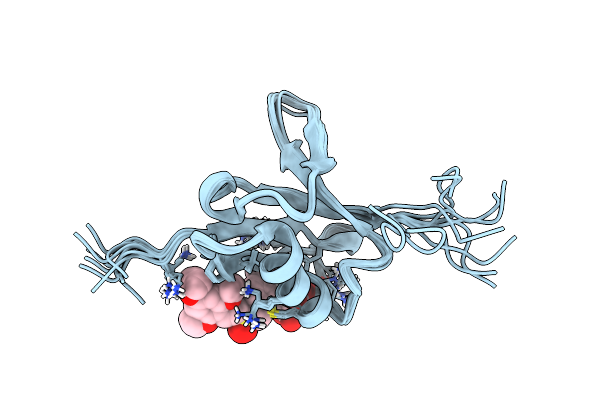 |
Organism: Homo sapiens
Method: SOLUTION NMR Release Date: 2016-05-04 Classification: PROTEIN BINDING Ligands: 6FS |
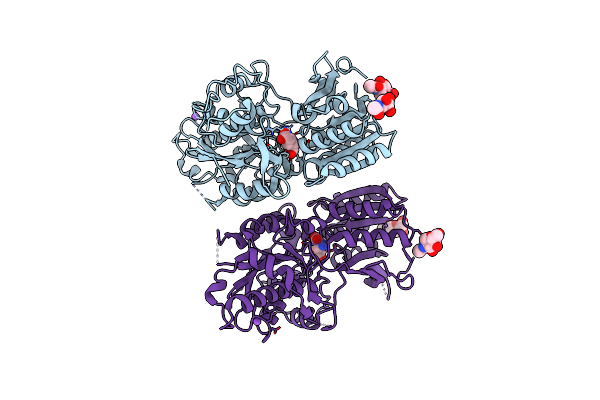 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.69 Å Release Date: 2015-09-09 Classification: SIGNALING PROTEIN Ligands: GLU, CL, NA, NAG |

