Search Count: 8
 |
Organism: Rattus norvegicus
Method: ELECTRON MICROSCOPY Release Date: 2022-11-23 Classification: MEMBRANE PROTEIN Ligands: ZN, CA, PLX |
 |
Structure Of The Full-Length Ip3R1 Channel Determined In The Presence Of Calcium/Ip3/Atp
Organism: Rattus norvegicus
Method: ELECTRON MICROSCOPY Release Date: 2022-11-23 Classification: MEMBRANE PROTEIN Ligands: ZN, ATP, I3P, CA, PLX |
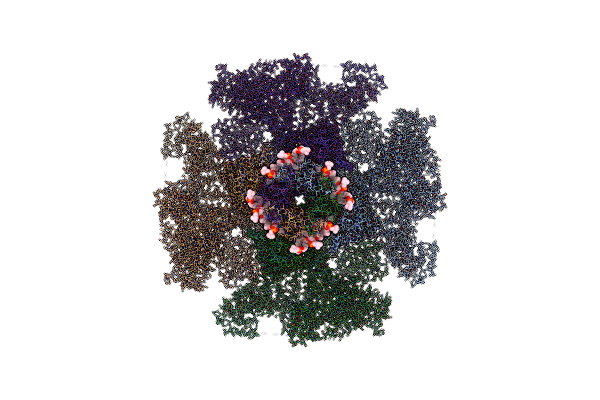 |
Structure Of Full-Length Ip3R1 Channel Reconstituted Into Lipid Nanodisc In The Apo-State
Organism: Rattus norvegicus
Method: ELECTRON MICROSCOPY Release Date: 2021-06-02 Classification: MEMBRANE PROTEIN Ligands: PLX, ZN |
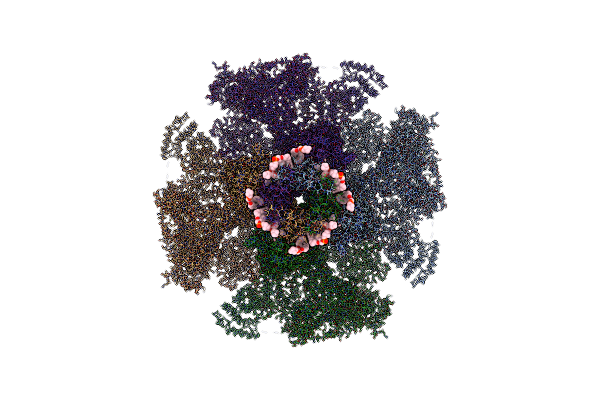 |
Structure Of Full-Length Ip3R1 Channel Solubilized In Lnmg & Lipid In The Apo-State
Organism: Rattus norvegicus
Method: ELECTRON MICROSCOPY Release Date: 2021-06-02 Classification: MEMBRANE PROTEIN Ligands: PLX, ZN |
 |
Organism: Rattus norvegicus
Method: ELECTRON MICROSCOPY Release Date: 2018-12-05 Classification: MEMBRANE PROTEIN Ligands: JYP |
 |
Organism: Rattus norvegicus
Method: ELECTRON MICROSCOPY Release Date: 2018-12-05 Classification: MEMBRANE PROTEIN |
 |
Structure Of Full-Length Ip3R1 Channel In The Apo-State Determined By Single Particle Cryo-Em
Organism: Rattus norvegicus
Method: ELECTRON MICROSCOPY Release Date: 2015-10-07 Classification: TRANSPORT PROTEIN |
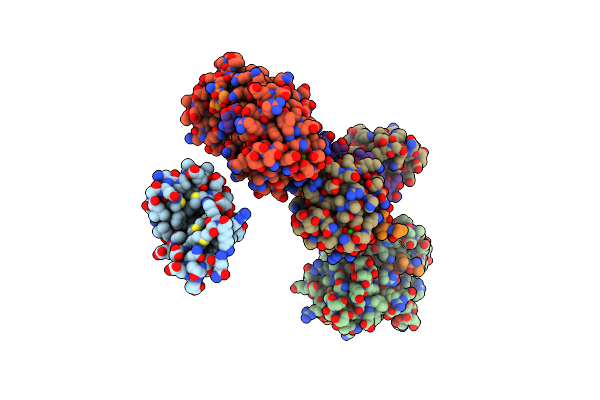 |
Crystal Structure Of The Calmodulin-Bound Cav1.2 C-Terminal Regulatory Domain Dimer
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2009-03-03 Classification: METAL BINDING PROTEIN Ligands: CA |

