Search Count: 30
 |
Cryo-Em Structure Of Human Exon-Defined Spliceosome In The Late Pre-B State.
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2024-05-01 Classification: SPLICING Ligands: IHP, MG, GTP, ZN |
 |
Cryo-Em Structure Of Human Exon-Defined Spliceosome In The Mature Pre-B State.
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2024-05-01 Classification: SPLICING Ligands: IHP, MG, GTP, ZN |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2024-05-01 Classification: SPLICING Ligands: IHP, GTP, MG, ZN |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2024-05-01 Classification: SPLICING Ligands: IHP, GTP, MG, ZN |
 |
Cryo-Em Structure Of The Human Minor Pre-B Complex (Pre-Precatalytic Spliceosome) U11 And Tri-Snrnp Part
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2024-03-20 Classification: SPLICING/RNA Ligands: MG, IHP, GTP, ZN |
 |
Cryo-Em Structure Of The Human Minor Pre-B Complex (Pre-Precatalytic Spliceosome) U12 Snrnp Part
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2024-03-13 Classification: SPLICING/RNA Ligands: ZN |
 |
Organism: Rattus norvegicus, Gallus gallus, Bos taurus
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2022-11-09 Classification: CELL CYCLE Ligands: GTP, MG, CA, GDP, MES, LNU, IMD, ACP |
 |
Organism: Rattus norvegicus, Gallus gallus, Bos taurus
Method: X-RAY DIFFRACTION Resolution:2.29 Å Release Date: 2022-11-09 Classification: CELL CYCLE Ligands: GTP, MG, CA, GDP, MES, LO9, ACP |
 |
Structural And Functional Characterization Of Bovine G1P[5] Rotavirus Vp8* Protein
Organism: Rotavirus a
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2021-12-29 Classification: VIRAL PROTEIN |
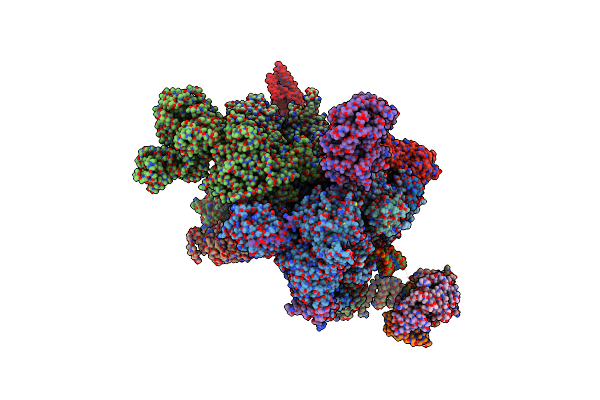 |
Cryo-Em Structure Of The Activated Human Minor Spliceosome (Minor Bact Complex)
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2021-03-31 Classification: SPLICING Ligands: IHP, GTP, MG, K, G5J, ZN |
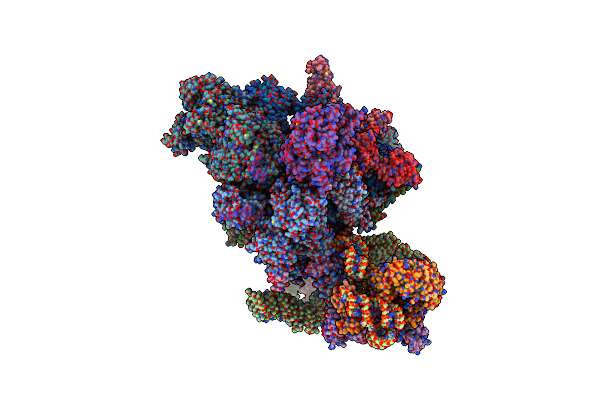 |
Cryo-Em Structure Of The Activated Spliceosome (Bact Complex) At An Atomic Resolution Of 2.5 Angstrom
Organism: Saccharomyces cerevisiae, Saccharomyces cerevisiae (strain atcc 204508 / s288c)
Method: ELECTRON MICROSCOPY Release Date: 2021-03-17 Classification: SPLICING Ligands: IHP, GTP, MG, CA, ZN |
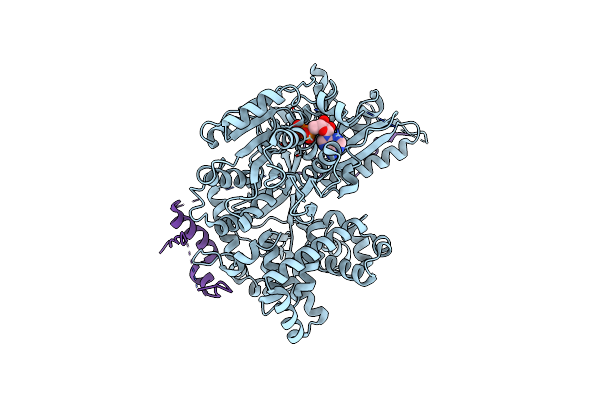 |
Organism: Saccharomyces cerevisiae
Method: ELECTRON MICROSCOPY Release Date: 2021-01-06 Classification: SPLICING Ligands: ADP, MG |
 |
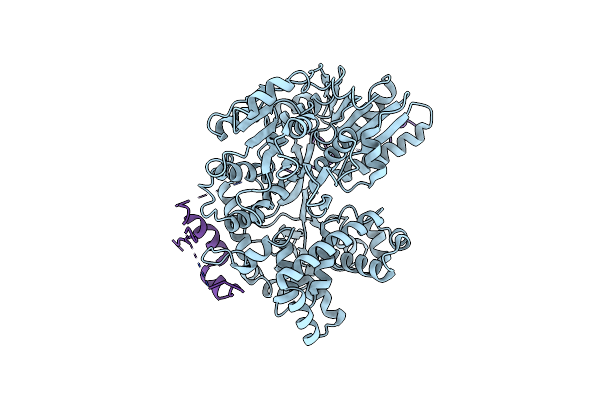
Organism: Saccharomyces cerevisiae
Method: ELECTRON MICROSCOPY Release Date: 2021-01-06 Classification: SPLICING |
 |
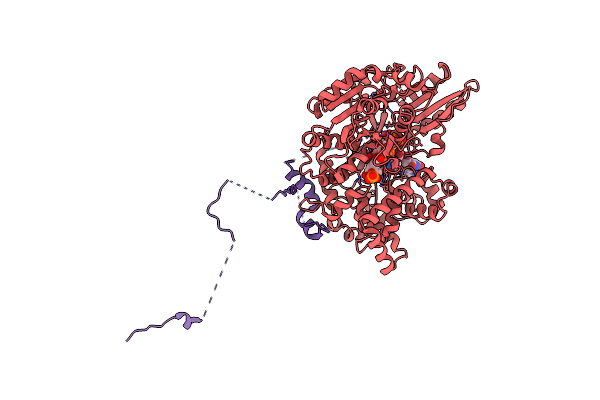
Cryo-Em Structure Of The Deah-Box Helicase Prp2 In Complex With Its Coactivator Spp2
Organism: Saccharomyces cerevisiae
Method: ELECTRON MICROSCOPY Release Date: 2021-01-06 Classification: SPLICING |
 |
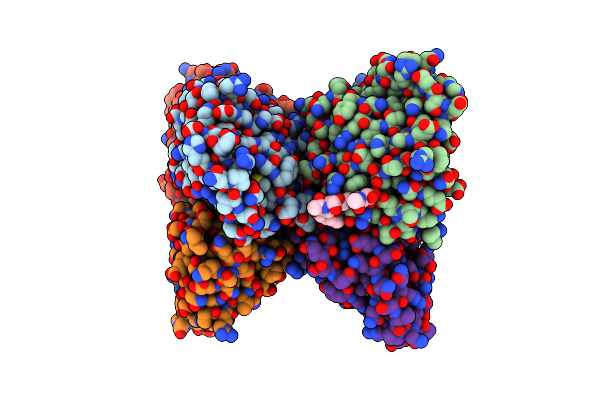
Cryo-Em Structure Of The Pre-Mrna-Loaded Deah-Box Atpase/Helicase Prp2 In Complex With Spp2
Organism: Saccharomyces cerevisiae
Method: ELECTRON MICROSCOPY Release Date: 2021-01-06 Classification: SPLICING |
 |
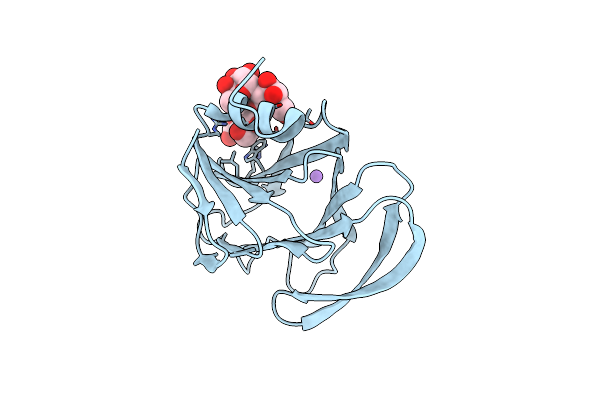
Structural Basis Of Glycan Recognition In Globally Predominant Human P[8] Rotavirus
Organism: Rotavirus a
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2019-10-09 Classification: VIRAL PROTEIN |
 |
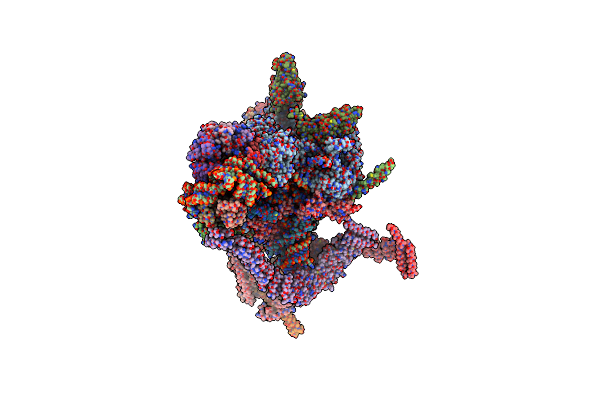
Structural Basis Of Glycan Recognition In Globally Predominant Human P[8] Rotavirus
Organism: Rotavirus a
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2019-10-09 Classification: VIRAL PROTEIN |
 |
Cryo-Em Structure Of The Yeast B*-A2 Complex At An Average Resolution Of 3.2 Angstrom
Organism: Saccharomyces cerevisiae (strain atcc 204508 / s288c), Saccharomyces cerevisiae s288c
Method: ELECTRON MICROSCOPY Release Date: 2019-04-24 Classification: SPLICING Ligands: IHP, GTP, MG, ZN |
 |
Cryo-Em Structure Of The Yeast B*-A1 Complex At An Average Resolution Of 3.6 Angstrom
Organism: Saccharomyces cerevisiae s288c, Saccharomyces cerevisiae (strain atcc 204508 / s288c)
Method: ELECTRON MICROSCOPY Release Date: 2019-04-24 Classification: SPLICING Ligands: IHP, GTP, MG, ZN |
 |
Cryo-Em Structure Of The Yeast B*-B1 Complex At An Average Resolution Of 3.86 Angstrom
Organism: Saccharomyces cerevisiae s288c
Method: ELECTRON MICROSCOPY Release Date: 2019-04-24 Classification: SPLICING Ligands: IHP, GTP, MG, ZN |

