Search Count: 8
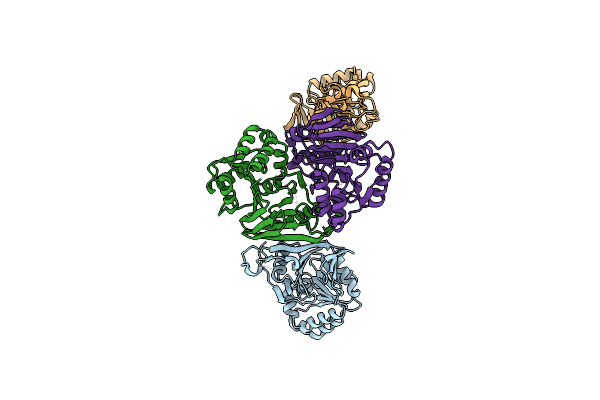 |
Organism: Pseudobacteroides cellulosolvens
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2022-08-10 Classification: HYDROLASE |
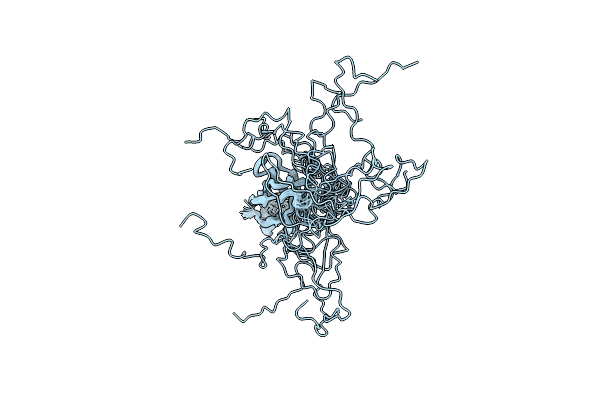 |
Solution Structure Of The Sensor Domain Of The Anti-Sigma Factor Rsgi4 In Pseudobacteroides Cellulosolvens
Organism: Pseudobacteroides cellulosolvens atcc 35603 = dsm 2933
Method: SOLUTION NMR Release Date: 2021-06-30 Classification: TRANSCRIPTION |
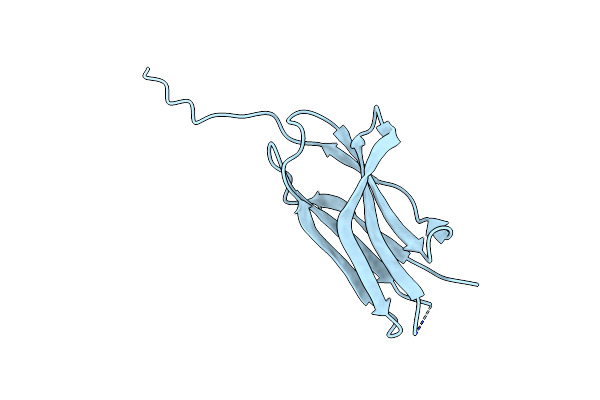 |
Structure Of The Sensor Domain (Long Construct) Of The Anti-Sigma Factor Rsgi4 In Pseudobacteroides Cellulosolvens
Organism: Pseudobacteroides cellulosolvens atcc 35603 = dsm 2933
Method: X-RAY DIFFRACTION Release Date: 2021-06-30 Classification: TRANSCRIPTION |
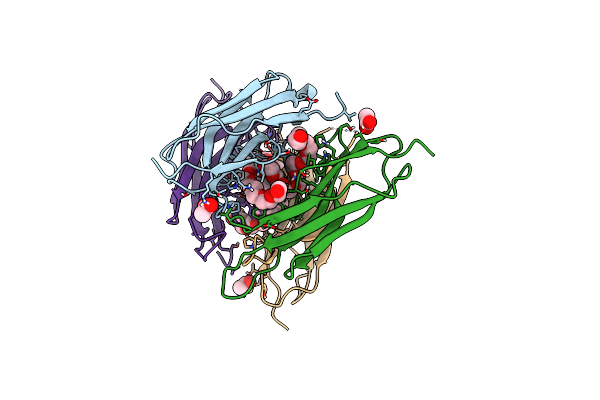 |
Structure Of The Sensor Domain (Short Construct) Of The Anti-Sigma Factor Rsgi4 In Pseudobacteroides Cellulosolvens
Organism: Pseudobacteroides cellulosolvens atcc 35603 = dsm 2933
Method: X-RAY DIFFRACTION Release Date: 2021-06-30 Classification: TRANSCRIPTION Ligands: ACT, PEU |
 |
The Crystal Structure Of The Seventh Scab Type I Cohesin From Pseudobacteroides Cellulosolvens
Organism: Bacteroides cellulosolvens
Method: X-RAY DIFFRACTION Resolution:1.84 Å Release Date: 2015-05-06 Classification: STRUCTURAL PROTEIN |
 |
Organism: Bacteroides cellulosolvens
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2012-01-11 Classification: STRUCTRUAL PROTEIN/HYDROLASE Ligands: CA |
 |
Structure Of A Scaffoldin Carbohydrate-Binding Module Family 3B From The Cellulosome Of Bacteroides Cellulosolvens: Structural Diversity And Implications For Carbohydrate Binding
Organism: Bacteroides cellulosolvens
Method: X-RAY DIFFRACTION Resolution:1.83 Å Release Date: 2011-04-06 Classification: SUGAR BINDING PROTEIN Ligands: NO3 |
 |
Crystal Structure Analysis Of Type Ii Cohesin A11 From Bacteroides Cellulosolvens
Organism: Bacteroides cellulosolvens
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2005-04-26 Classification: STRUCTURAL PROTEIN Ligands: EDO, MOH |

