Search Count: 4,019
All
Selected
 |
X-Ray Structure Of The Bacteroides Fragilis Nramp/Mnth Divalent Transition Metal Transporter Wt In An Inward-Open, State
Organism: Bacteroides fragilis
Method: X-RAY DIFFRACTION Resolution:2.48 Å Release Date: 2026-03-04 Classification: METAL TRANSPORT Ligands: OLC, PEG |
 |
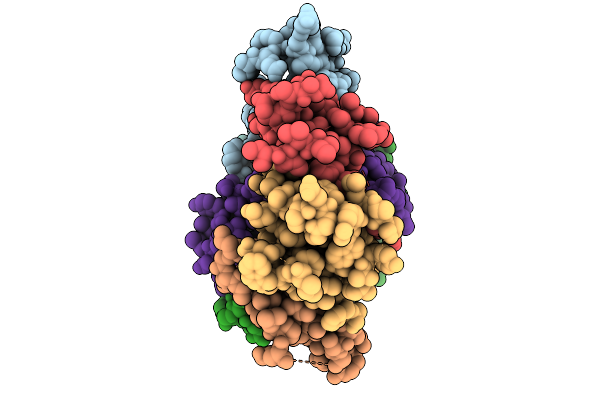
X-Ray Structure Of The Bacteroides Fragilis Nramp/Mnth Divalent Transition Metal Transporter Wt In An Inward-Open, Manganese-Bound State
Organism: Bacteroides fragilis
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2026-03-04 Classification: METAL TRANSPORT Ligands: MN, OLC, PEG |
 |
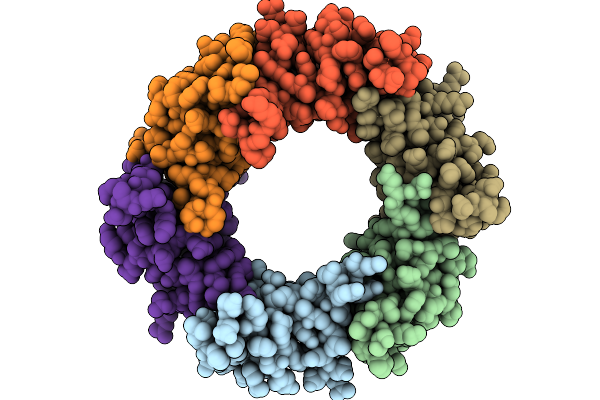
Structure Of The Bacteroides Fragilis Nctc9343 T6Ss Hcp2-Hcp3 Heterohexamer In Complex With The Effector Bte1
Organism: Bacteroides fragilis nctc 9343
Method: ELECTRON MICROSCOPY Resolution:3.30 Å Release Date: 2026-01-21 Classification: STRUCTURAL PROTEIN |
 |
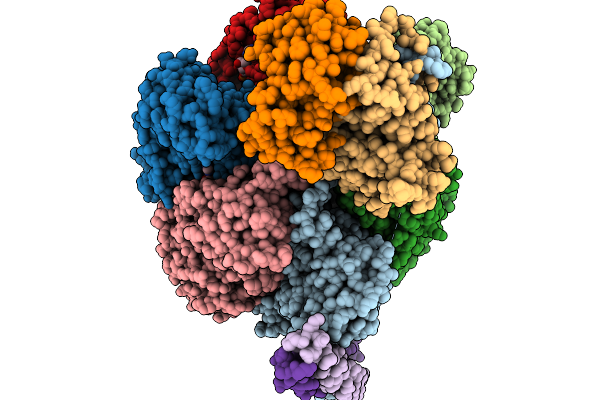
Organism: Bacteroides fragilis nctc 9343
Method: ELECTRON MICROSCOPY Resolution:3.11 Å Release Date: 2026-01-21 Classification: STRUCTURAL PROTEIN |
 |
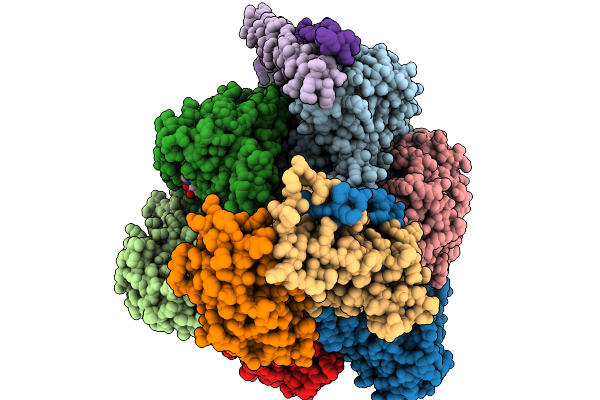
Cryo-Em Structure Of F-Atp Synthase From Mycobacteroides Abscessus (Rotational State 1)
Organism: Mycobacteroides abscessus subsp. abscessus
Method: ELECTRON MICROSCOPY Resolution:2.94 Å Release Date: 2026-01-14 Classification: HYDROLASE Ligands: ATP, MG, ADP |
 |
Cryo-Em Structure Of F-Atp Synthase From Mycobacteroides Abscessus (Rotational State 2)
Organism: Mycobacteroides abscessus subsp. abscessus
Method: ELECTRON MICROSCOPY Resolution:3.41 Å Release Date: 2026-01-14 Classification: HYDROLASE Ligands: ATP, MG, ADP |
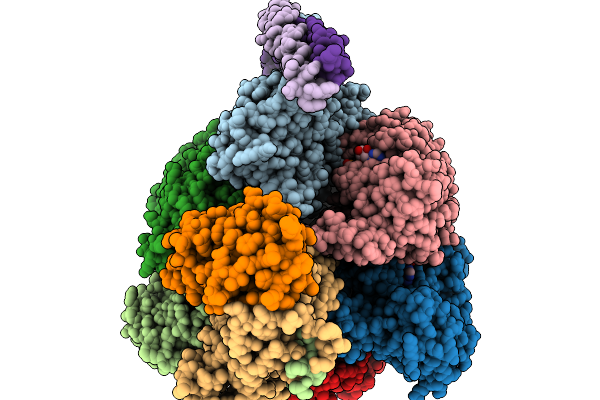 |
Cryo-Em Structure Of F-Atp Synthase From Mycobacteroides Abscessus (Rotational State 3)
Organism: Mycobacteroides abscessus subsp. abscessus
Method: ELECTRON MICROSCOPY Resolution:2.79 Å Release Date: 2026-01-14 Classification: HYDROLASE Ligands: ATP, MG, ADP |
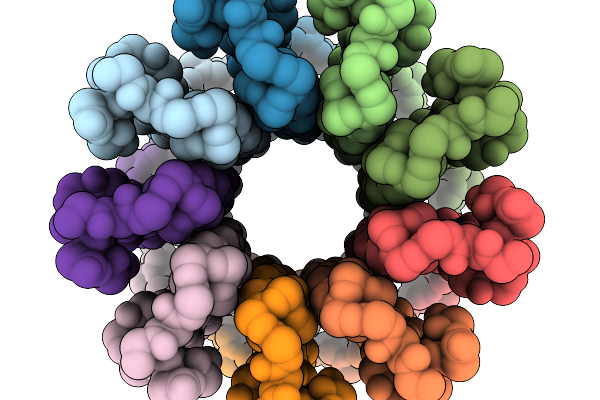 |
Cryo-Em Structure Of F-Atp Synthase C-Ring From Mycobacteroides Abscessus (Backbone)
Organism: Mycobacteroides abscessus subsp. abscessus
Method: ELECTRON MICROSCOPY Resolution:5.61 Å Release Date: 2026-01-14 Classification: HYDROLASE |
 |
Organism: Mycobacteroides abscessus
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2026-01-07 Classification: OXIDOREDUCTASE Ligands: FAD |
 |
Organism: Bacteroides fragilis
Method: X-RAY DIFFRACTION Resolution:1.94 Å Release Date: 2026-01-07 Classification: TOXIN Ligands: ZN |
 |
Organism: Bacteroides fragilis
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2026-01-07 Classification: TOXIN Ligands: ZN |
 |
Organism: Homo sapiens, Bacteroides uniformis atcc 8492
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2025-12-03 Classification: BIOSYNTHETIC PROTEIN Ligands: ACT, NA |
 |
Organism: Bacteroides uniformis atcc 8492
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2025-12-03 Classification: BIOSYNTHETIC PROTEIN Ligands: IPA |
 |
Organism: Homo sapiens, Bacteroides uniformis atcc 8492
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2025-12-03 Classification: BIOSYNTHETIC PROTEIN Ligands: CIT, PEG |
 |
Crystal Structure Of The Nte Domain Of Suscdex (Bt3090) Dextran Transporter
Organism: Bacteroides thetaiotaomicron vpi-5482
Method: X-RAY DIFFRACTION Resolution:1.40 Å Release Date: 2025-11-26 Classification: TRANSPORT PROTEIN Ligands: NI, SCN, EDO, PEG, PGE |
 |
Organism: Bacteroides fragilis
Method: ELECTRON MICROSCOPY Release Date: 2025-11-26 Classification: IMMUNE SYSTEM/RNA Ligands: MN |
 |
Organism: Bacteroides fragilis
Method: ELECTRON MICROSCOPY Release Date: 2025-11-26 Classification: IMMUNE SYSTEM/RNA Ligands: ATP, SAM, MN |
 |
Organism: Bacteroides fragilis
Method: ELECTRON MICROSCOPY Release Date: 2025-11-26 Classification: IMMUNE SYSTEM/RNA Ligands: ATP, SAM, MN |
 |
Organism: Bacteroides fragilis
Method: ELECTRON MICROSCOPY Release Date: 2025-11-26 Classification: IMMUNE SYSTEM/RNA Ligands: ATP, SAM, MN |
 |
Organism: Bacteroides fragilis
Method: X-RAY DIFFRACTION Resolution:2.38 Å Release Date: 2025-11-26 Classification: IMMUNE SYSTEM Ligands: MN |

