Search Count: 386
 |
 |
Organism: Caloramator australicus rc3, Unidentified
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2025-12-10 Classification: METAL BINDING PROTEIN Ligands: ZN, NA, EPE, MG |
 |
Cryo-Em Map Of Msmeg_3496 In Complex With Acpm, Size Exclusion Chromatography Peak1
Organism: Mycolicibacterium smegmatis mc2 155
Method: ELECTRON MICROSCOPY Release Date: 2025-12-10 Classification: MEMBRANE PROTEIN |
 |
Cryo-Em Map Of Msmeg_3496 In Complex With Acpm, Size Exclusion Chromatography Peak2
Organism: Mycolicibacterium smegmatis mc2 155
Method: ELECTRON MICROSCOPY Release Date: 2025-12-10 Classification: MEMBRANE PROTEIN |
 |
Organism: Mycobacterium tuberculosis (strain atcc 25618 / h37rv), Mycolicibacterium smegmatis mc2 155
Method: ELECTRON MICROSCOPY Release Date: 2025-12-10 Classification: MEMBRANE PROTEIN |
 |
Organism: Mycobacterium tuberculosis, Aequorea victoria, Mycolicibacterium smegmatis mc2 155
Method: ELECTRON MICROSCOPY Resolution:3.20 Å Release Date: 2025-10-08 Classification: MEMBRANE PROTEIN |
 |
Organism: Mycobacterium tuberculosis h37rv, Mycobacterium tuberculosis, Mycolicibacterium smegmatis mc2 155
Method: ELECTRON MICROSCOPY Resolution:3.30 Å Release Date: 2025-08-13 Classification: MEMBRANE PROTEIN Ligands: L9Q |
 |
Organism: Mycobacterium tuberculosis, Aequorea victoria, Mycolicibacterium smegmatis mc2 155
Method: ELECTRON MICROSCOPY Resolution:3.20 Å Release Date: 2025-08-13 Classification: MEMBRANE PROTEIN Ligands: L9Q |
 |
Organism: Mycobacterium tuberculosis
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2025-07-30 Classification: BIOSYNTHETIC PROTEIN Ligands: ALA, ZN |
 |
Organism: Mycolicibacterium smegmatis
Method: ELECTRON MICROSCOPY Release Date: 2025-07-30 Classification: MEMBRANE PROTEIN Ligands: PNS |
 |
Organism: Mycolicibacterium smegmatis
Method: ELECTRON MICROSCOPY Release Date: 2025-07-30 Classification: MEMBRANE PROTEIN Ligands: PNS |
 |
Cluster Of Bipartite Complex Of Mmpl5-Acpm From Mycolicibacterium Smegmatis
Organism: Mycolicibacterium smegmatis
Method: ELECTRON MICROSCOPY Release Date: 2025-07-30 Classification: MEMBRANE PROTEIN Ligands: PNS |
 |
Imidazole Glycerol Phosphate Dehydratase From Mycobacterium Tuberculosis, Apo Structure
Organism: Mycobacterium tuberculosis
Method: ELECTRON MICROSCOPY Resolution:3.10 Å Release Date: 2025-06-18 Classification: LYASE Ligands: MN |
 |
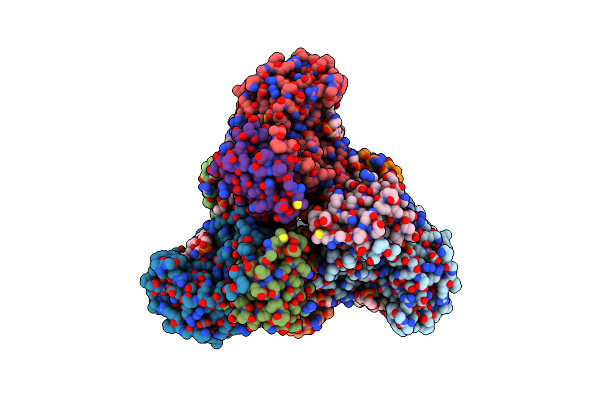
Imidazole Glycerol Phosphate Dehydratase From Mycobacterium Tuberculosis, In Complex With Aminotriazole
Organism: Mycobacterium tuberculosis
Method: ELECTRON MICROSCOPY Resolution:2.80 Å Release Date: 2025-06-18 Classification: LYASE Ligands: MN, 3TR |
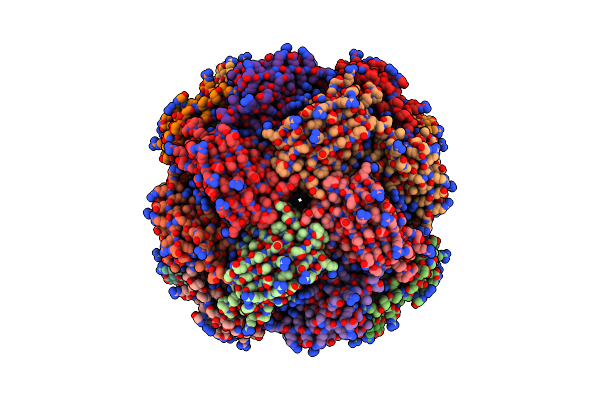 |
Imidazole Glycerol Phosphate Dehydratase From Mycobacterium Tuberculosis, Apo Structure
Organism: Mycobacterium tuberculosis
Method: ELECTRON MICROSCOPY Resolution:2.20 Å Release Date: 2025-06-18 Classification: LYASE Ligands: MN |
 |
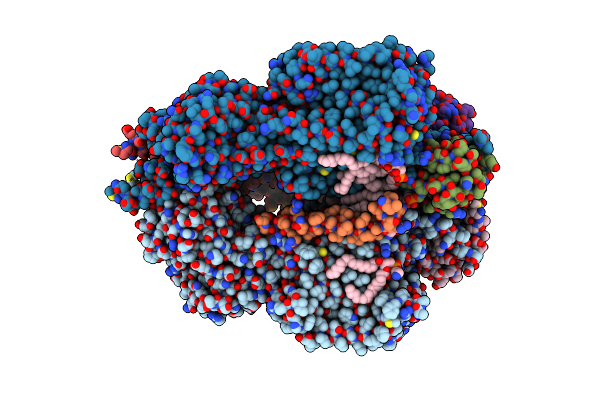
Cryo-Em Structure Of The Efflux Transporter Mmpl5/Mmps5 From Mycobacterium Tuberculosis, C3 Symmetry
Organism: Mycobacterium tuberculosis h37rv, Mycolicibacterium smegmatis mc2 155
Method: ELECTRON MICROSCOPY Release Date: 2025-05-21 Classification: MEMBRANE PROTEIN Ligands: CDL, PEV, PN7 |
 |
Cryo-Em Structure Of The Efflux Transporter Mmpl5/Mmps5 From Mycobacterium Tuberculosis, C1 Symmetry
Organism: Mycobacterium tuberculosis h37rv, Mycolicibacterium smegmatis mc2 155
Method: ELECTRON MICROSCOPY Release Date: 2025-05-21 Classification: MEMBRANE PROTEIN Ligands: CDL, PEV, PN7 |
 |
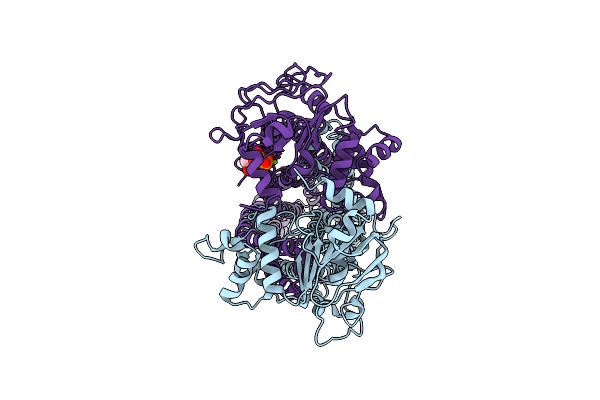
Cryo-Em Structure Of Msrv1273C/72C From Mycobacterium Smegmatis In The Atp|Adp-Bound Ifasym-2 State
Organism: Mycolicibacterium smegmatis mc2 155
Method: ELECTRON MICROSCOPY Release Date: 2025-05-14 Classification: TRANSPORT PROTEIN Ligands: ATP, MG, ADP |
 |
Cryo-Em Structure Of Msrv1273C/72C From Mycobacterium Smegmatis In The Adp-Bound Ifasym-3 State (Atp 37 Degrees C Treated
Organism: Mycolicibacterium smegmatis mc2 155
Method: ELECTRON MICROSCOPY Release Date: 2025-05-14 Classification: TRANSPORT PROTEIN Ligands: ADP, MG |
 |
Cryo-Em Structure Of Msrv1273C/72C From Mycobacterium Smegmatis In The Adp-Bound Ifasym-3 State (Adp 4 Degrees C Treated)
Organism: Mycolicibacterium smegmatis mc2 155
Method: ELECTRON MICROSCOPY Release Date: 2025-05-14 Classification: TRANSPORT PROTEIN Ligands: ADP, MG |

