Search Count: 48
 |
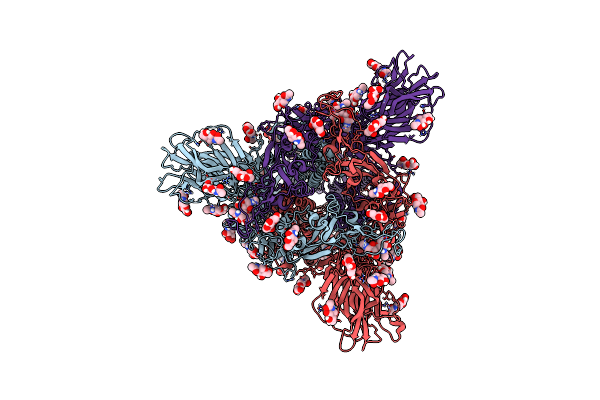
Cryo-Em Structure Of Eu-Hedgehogcov (Erinaceus/Vmc/Deu/2012) S-Trimer In A Locked-2 Conformation
Organism: Betacoronavirus erinaceus/vmc/deu/2012, Enterobacteria phage t4 (bacteriophage t4)
Method: ELECTRON MICROSCOPY Release Date: 2025-06-18 Classification: VIRAL PROTEIN Ligands: NAG, FOL, EIC |
 |
Organism: Severe acute respiratory syndrome coronavirus, Enterobacteria phage t4 (bacteriophage t4)
Method: ELECTRON MICROSCOPY Release Date: 2023-08-16 Classification: BIOSYNTHETIC PROTEIN Ligands: NAG |
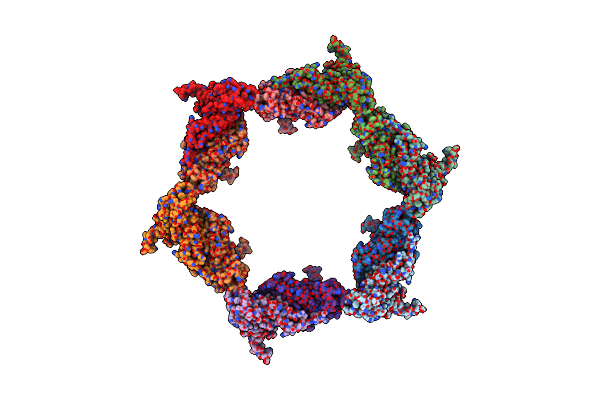 |
Fitting Of The Gp6 Crystal Structure Into 3D Cryo-Em Reconstruction Of Bacteriophage T4 Star-Shaped Baseplate
Organism: Bacteriophage t4
Method: ELECTRON MICROSCOPY Release Date: 2009-05-19 Classification: VIRAL PROTEIN |
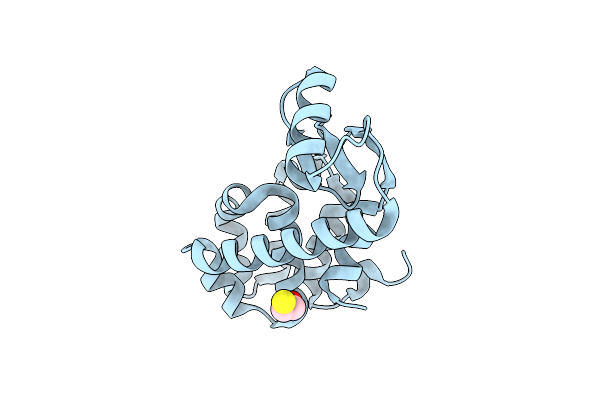 |
Contributions Of All 20 Amino Acids At Site 96 To The Stability And Structure Of T4 Lysozyme
Organism: Bacteriophage t4
Method: X-RAY DIFFRACTION Resolution:1.77 Å Release Date: 2009-02-17 Classification: HYDROLASE Ligands: CL, BME |
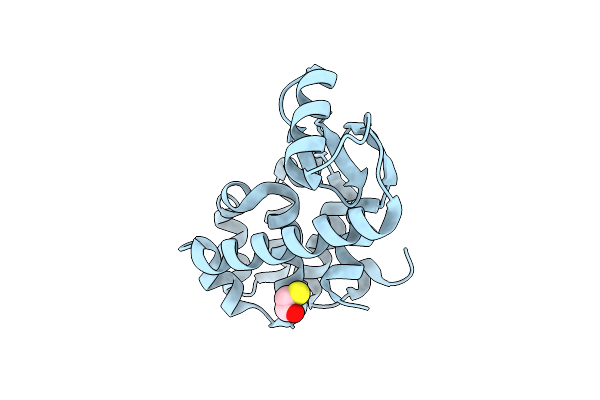 |
Organism: Bacteriophage t4
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2009-02-17 Classification: HYDROLASE Ligands: CL, BME |
 |
Organism: Bacteriophage t4
Method: X-RAY DIFFRACTION Resolution:1.67 Å Release Date: 2009-02-17 Classification: HYDROLASE Ligands: CL, BME |
 |
Organism: Bacteriophage t4
Method: X-RAY DIFFRACTION Resolution:1.99 Å Release Date: 2009-02-17 Classification: HYDROLASE Ligands: CL, BME |
 |
Organism: Bacteriophage t4
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2009-02-17 Classification: HYDROLASE Ligands: CL, BME |
 |
Organism: Bacteriophage t4
Method: X-RAY DIFFRACTION Resolution:1.68 Å Release Date: 2009-02-17 Classification: HYDROLASE Ligands: CL, BME |
 |
Bacteriophage T4 Lysozyme Mutant D89A In Wildtype Background At Room Temperature
Organism: Bacteriophage t4
Method: X-RAY DIFFRACTION Resolution:1.84 Å Release Date: 2009-02-17 Classification: HYDROLASE Ligands: CL, BME |
 |
Contribution Of All 20 Amino Acids At Site 96 To The Stability And Structure Of T4 Lysozyme
Organism: Bacteriophage t4
Method: X-RAY DIFFRACTION Resolution:1.64 Å Release Date: 2009-02-17 Classification: HYDROLASE Ligands: K, CL, BME |
 |
Contributions Of All 20 Amino Acids At Site 96 To Stability And Structure Of T4 Lysozyme
Organism: Bacteriophage t4
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2009-02-17 Classification: HYDROLASE Ligands: CL, BME |
 |
Contributions Of All 20 Amino Acids At Site 96 To The Stability And Structure Of T4 Lysozyme
Organism: Bacteriophage t4
Method: X-RAY DIFFRACTION Resolution:1.68 Å Release Date: 2009-02-17 Classification: HYDROLASE Ligands: CL, K, BME |
 |
Bacteriophage T4 Lysozyme Mutant R96V In Wildtype Background At Low Temperature
Organism: Bacteriophage t4
Method: X-RAY DIFFRACTION Resolution:1.87 Å Release Date: 2009-02-17 Classification: HYDROLASE Ligands: HEZ, MPD, SO4 |
 |
Contributions Of All 20 Amino Acids At Site 96 To The Stability And Structure Of T4 Lysozyme
Organism: Bacteriophage t4
Method: X-RAY DIFFRACTION Resolution:1.68 Å Release Date: 2009-02-17 Classification: HYDROLASE Ligands: CL, K, BME |
 |
Organism: Bacteriophage t4
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2009-02-17 Classification: HYDROLASE Ligands: CL, HED |
 |
Contributions Of All 20 Amino Acids At Site 96 To The Stability And Structure Of T4 Lysozyme
Organism: Bacteriophage t4
Method: X-RAY DIFFRACTION Resolution:1.63 Å Release Date: 2009-02-17 Classification: HYDROLASE Ligands: CL, BME |
 |
Contributions Of All 20 Amino Acids At Site 96 To The Stability And Structure Of T4 Lysozyme
Organism: Bacteriophage t4
Method: X-RAY DIFFRACTION Resolution:1.73 Å Release Date: 2009-02-17 Classification: HYDROLASE Ligands: CL, BME |
 |
Organism: Bacteriophage t4
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2009-01-27 Classification: HYDROLASE Ligands: PO4, CL, HED, BNZ |
 |
Organism: Bacteriophage t4
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2009-01-13 Classification: HYDROLASE Ligands: PO4, NA |

