Search Count: 30
 |
The Crystal Structure Of Domain-Swapped Dimer Q108K:T51D:A28C:L36C:F57:H:R58 Mutant Of Hcrbpii With A Histidine Insertion In The Hinge Loop Region At 1.96 Angstrom Resolution
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.96 Å Release Date: 2023-10-18 Classification: LIPID BINDING PROTEIN |
 |
The Crystal Structure Of Domain-Swapped Dimer Q108K:T51D:A28C:L36C:F57:H:H:H:R58 Mutant Of Hcrbpii With Histidine Insertion In The Hinge Loop Region At 1.92 Angstrom Resolution
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.92 Å Release Date: 2023-10-18 Classification: LIPID BINDING PROTEIN |
 |
The Crystal Structure Of Apo Domain-Swapped Dimer F57:H:H:H:H:H:H:R58 Mutant Of Hcrbpii With Histidine Insertion In The Hinge Loop Region At 2.5 Angstrom Resolution
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.52 Å Release Date: 2023-10-18 Classification: LIPID BINDING PROTEIN |
 |
The Crystal Structure Of Apo Domain-Swapped Dimer Q108K:T51D:A28Cl36C R58:H:H:H:N59 Hcrbpii With Histidine Insertion In The Hinge Loop Region At 2.19 Angstrom Resolution
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.19 Å Release Date: 2023-10-18 Classification: LIPID BINDING PROTEIN |
 |
The Crystal Structure Of Apo Monomer F57:H:H:H:H:H:H:R58 Mutant Of Hcrbpii With Histidine Insertion In The Hinge Loop Region At 1.1 Angstrom Resolution
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.08 Å Release Date: 2023-10-18 Classification: LIPID BINDING PROTEIN |
 |
The Crystal Structure Of Apo Monomer F57:H:H:H:R58 Hcrbpii With Histidine Insertion In The Hinge Loop Region At 1.3 Angstrom Resolution
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.32 Å Release Date: 2023-10-18 Classification: LIPID BINDING PROTEIN |
 |
The Crystal Structure Of Q108K:K40L:T51V:T53S:R58W:Y19W:A33W:L117E Mutant Of Hcrbpii Bound With Lizfluor
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.26 Å Release Date: 2022-01-12 Classification: LIPID BINDING PROTEIN Ligands: ACT, ZFG, GOL |
 |
The Crystal Structure Of Domain-Swapped Trimer Q108K:K40D:T53A:R58L:Q38F:Q4F Mutant Of Hcrbpii Bound With Lizfluor3 Chromophore Showing Excited State Intermolecular Proton Transfer
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.49 Å Release Date: 2022-01-12 Classification: LIPID BINDING PROTEIN Ligands: ZFP, ACT, GOL |
 |
The Crystal Structure Of Apo Domain-Swapped Dimer Q108K:K40D:T53A:R58L:Q38F:Q4F:F57H Variant Of Hcrbpii
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.67 Å Release Date: 2020-08-19 Classification: LIPID BINDING PROTEIN Ligands: ACT, GOL |
 |
The Crystal Structure Of Apo Domain-Swapped Trimer Q108K:T51D:A28C:I32C Of Hcrbpii
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2020-08-19 Classification: LIPID BINDING PROTEIN Ligands: GOL, 144, SO4 |
 |
The Crystal Structure Of Domain-Swapped Trimer Q108K:T51D Variant Of Hcrbpii
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.78 Å Release Date: 2020-08-19 Classification: LIPID BINDING PROTEIN Ligands: GOL, ACT |
 |
The Crystal Structure Of Apo Domain-Swapped Trimer Q108K:K40L:T51K Variant Of Hcrbpii
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.99 Å Release Date: 2020-08-19 Classification: LIPID BINDING PROTEIN Ligands: ACT, GOL |
 |
Crystal Structure Of The Apo Domain-Swapped Dimer Q108K:K40L:T51F Mutant Of Human Cellular Retinol Binding Protein Ii
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.97 Å Release Date: 2019-10-16 Classification: CYTOSOLIC PROTEIN Ligands: ACT |
 |
Crystal Structure Of The Apo Domain-Swapped Dimer Q108K:K40L:T51W Mutant Of Human Cellular Retinol Binding Protein Ii
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.26 Å Release Date: 2019-10-16 Classification: LIPID BINDING PROTEIN Ligands: HOH |
 |
Crystal Structure Of The Apo Domain-Swapped Dimer Q108K:T51D Mutant Of Human Cellular Retinol Binding Protein Ii
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2019-10-16 Classification: LIPID BINDING PROTEIN Ligands: ACT |
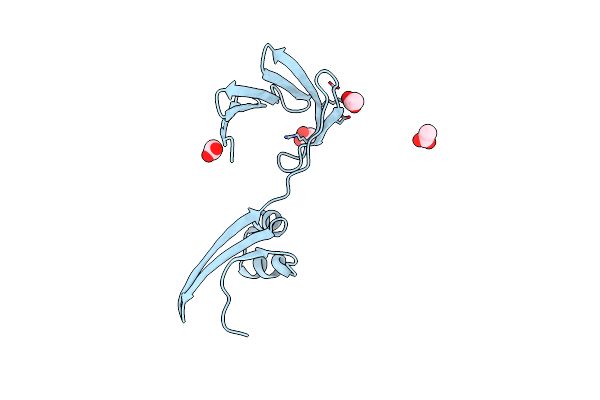 |
Crystal Structure Of The Apo Domain-Swapped Dimer Q108K:T51D:A28H Mutant Of Human Cellular Retinol Binding Protein Ii
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.99 Å Release Date: 2019-10-16 Classification: LIPID BINDING PROTEIN Ligands: ACT |
 |
Crystal Structure Of The Holo Retinal-Bound Domain-Swapped Dimer Q108K:T51D:A28C Mutant Of Human Cellular Retinol Binding Protein Ii
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2019-10-16 Classification: LIPID BINDING PROTEIN Ligands: ACT, GOL, RET |
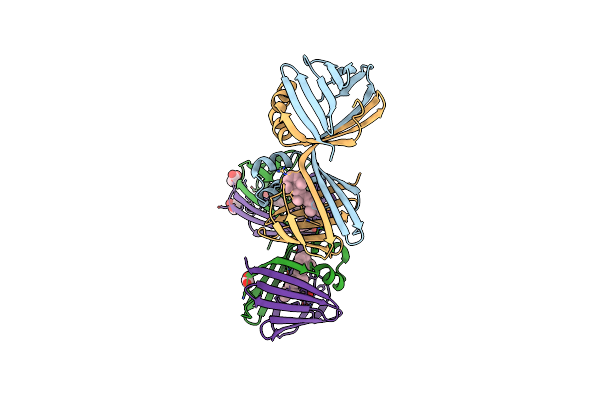 |
Crystal Structure Of Retinal-Bound Holo Q108K:K40L:T51W Domain-Swapped Dimer Of Human Cellular Retinol Binding Protein 2
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.11 Å Release Date: 2019-10-16 Classification: LIPID BINDING PROTEIN Ligands: RET, ACT, GOL |
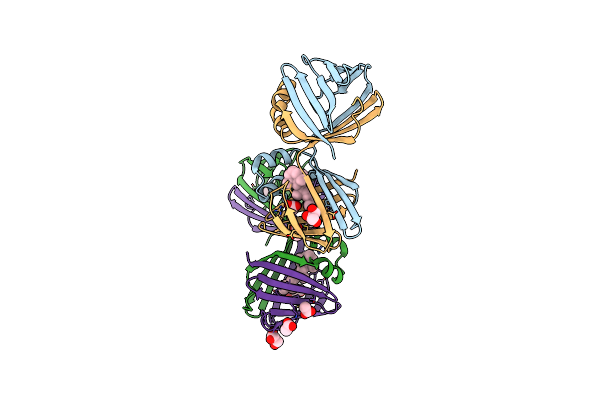 |
Crystal Structure Of The Holo Retinal-Bound Domain-Swapped Dimer Q108K:K40L:T51F Mutant Of Human Cellular Retinol Binding Protein Ii
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.15 Å Release Date: 2019-10-16 Classification: LIPID BINDING PROTEIN Ligands: GOL, ACT, RET |
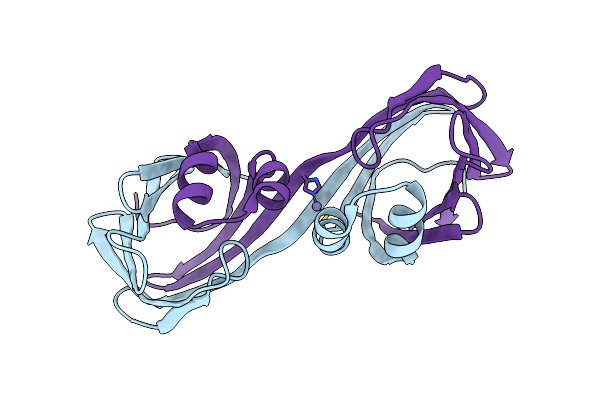 |
Crystal Structure Of The Zn-Bound Domain-Swapped Dimer Q108K:T51D:A28C:L36C:F57H Mutant Of Human Cellular Retinol Binding Protein Ii
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.64 Å Release Date: 2019-10-16 Classification: LIPID BINDING PROTEIN Ligands: ZN |

