Search Count: 7,574
All
Selected
 |
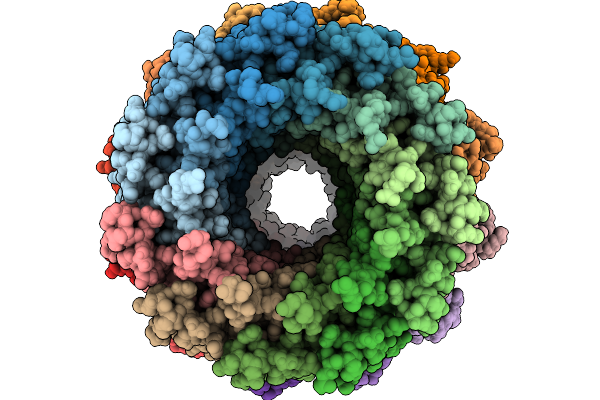
Organism: Escherichia coli k-12, Enterobacteria phage m
Method: ELECTRON MICROSCOPY Resolution:3.60 Å Release Date: 2026-02-11 Classification: TRANSPORT PROTEIN |
 |
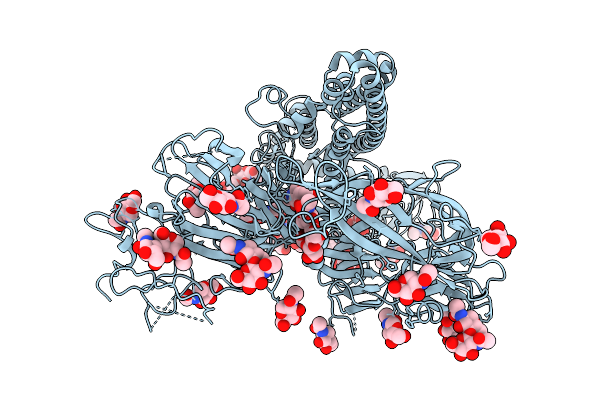
Cryo-Em Structure Of The Escherichia Phage Hk446 Rip1 In Complex With The Enterobacteria Phage T6 Small Terminase
Organism: Enterobacteria phage t6, Escherichia phage hk446
Method: ELECTRON MICROSCOPY Release Date: 2026-02-04 Classification: VIRAL PROTEIN |
 |
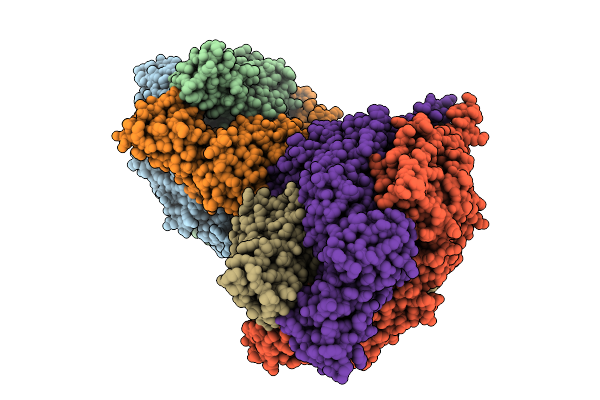
Organism: Feline infectious peritonitis virus (strain 79-1146), Enterobacteria phage t6
Method: ELECTRON MICROSCOPY Resolution:2.78 Å Release Date: 2026-01-28 Classification: PROTEIN BINDING Ligands: NAG |
 |
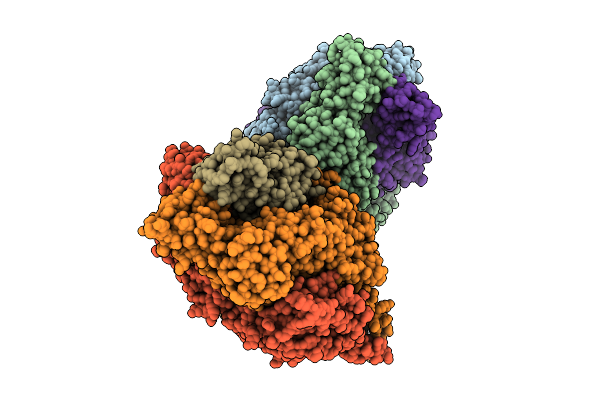
Organism: Human respiratory syncytial virus, Enterobacteria phage t6
Method: X-RAY DIFFRACTION Resolution:2.92 Å Release Date: 2025-12-17 Classification: VIRAL PROTEIN |
 |
Organism: Respiratory syncytial virus, Enterobacteria phage t6
Method: X-RAY DIFFRACTION Resolution:2.77 Å Release Date: 2025-12-17 Classification: VIRAL PROTEIN |
 |
Crystal Structure Of Phosphatidyl Inositol 4-Kinase Ii Beta In Complex With Hh5129
Organism: Homo sapiens, Enterobacteria phage t4
Method: X-RAY DIFFRACTION Resolution:2.25 Å Release Date: 2025-12-03 Classification: TRANSFERASE Ligands: A1IVA |
 |
Organism: Enterobacteria phage t4
Method: ELECTRON MICROSCOPY Release Date: 2025-11-26 Classification: ISOMERASE |
 |
Crystal Structure Of The Poly(Hexamethylene Adipamide) (Nylon66) Hydrolase Nyl50 In Its Apo Form At Room Temperature
Organism: Alphaproteobacteria bacterium
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2025-11-19 Classification: HYDROLASE |
 |
Organism: Rhodococcus sp. (in: high g+c gram-positive bacteria)
Method: X-RAY DIFFRACTION Resolution:2.04 Å Release Date: 2025-10-08 Classification: LIGASE Ligands: CL, A1JC1 |
 |
Crystal Structure Of Octameric Commd-Like Protein Mbk5214510 From Flavobacteriaceae Bacterium
Organism: Flavobacteriaceae bacterium
Method: X-RAY DIFFRACTION Resolution:2.58 Å Release Date: 2025-10-01 Classification: UNKNOWN FUNCTION |
 |
Cryo-Em Structure Of Octameric Commd-Like Protein Mbk5214510 From Flavobacteriaceae Bacterium
Organism: Flavobacteriaceae bacterium
Method: ELECTRON MICROSCOPY Release Date: 2025-10-01 Classification: UNKNOWN FUNCTION |
 |
Cryoem Structure Of Decameric Commd-Like Protein Mbk5214510 From Flavobacteriaceae Bacterium
Organism: Flavobacteriaceae bacterium
Method: ELECTRON MICROSCOPY Release Date: 2025-10-01 Classification: UNKNOWN FUNCTION |
 |
Organism: Escherichia coli, Enterobacteriaceae
Method: ELECTRON MICROSCOPY Resolution:2.90 Å Release Date: 2025-09-24 Classification: MEMBRANE PROTEIN |
 |
Organism: Escherichia coli, Enterobacteria phage m
Method: ELECTRON MICROSCOPY Release Date: 2025-09-10 Classification: VIRAL PROTEIN |
 |
Cryo-Em Structure Of Candidatus Saccharibacterium Phosphoketolase Complexed With Thiamine Diphosphate
Organism: Candidatus saccharibacteria bacterium
Method: ELECTRON MICROSCOPY Release Date: 2025-08-06 Classification: LYASE Ligands: TPP |
 |
Cryo-Em Structure Of Candidatus Saccharibacterium Phosphoketolase Complexed With 2-Acetyl-Thiamine Diphosphate
Organism: Candidatus saccharibacteria bacterium
Method: ELECTRON MICROSCOPY Release Date: 2025-08-06 Classification: LYASE Ligands: HTL |
 |
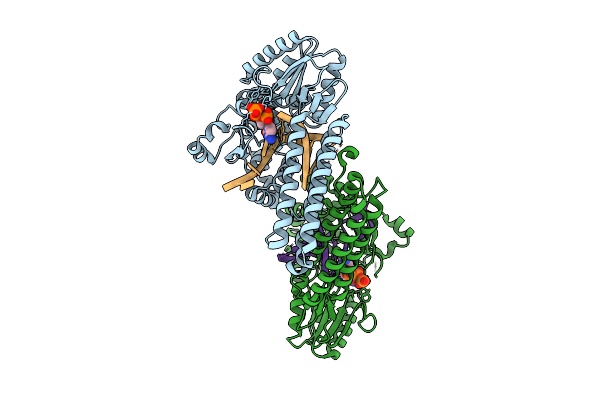
Organism: Gammaproteobacteria
Method: X-RAY DIFFRACTION Resolution:2.61 Å Release Date: 2025-08-06 Classification: DNA BINDING PROTEIN Ligands: URE |
 |
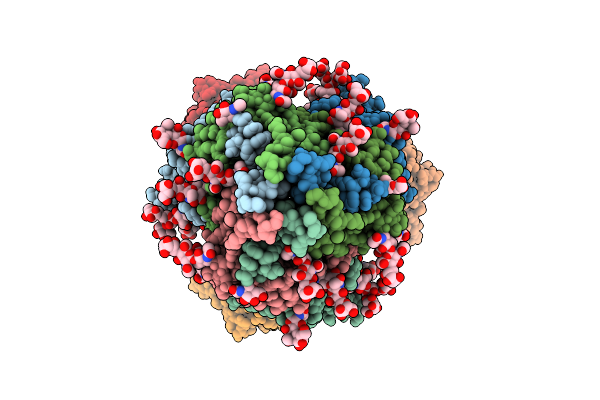
Organism: Gammaproteobacteria, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.77 Å Release Date: 2025-07-30 Classification: DNA BINDING PROTEIN Ligands: MN, DCP |
 |
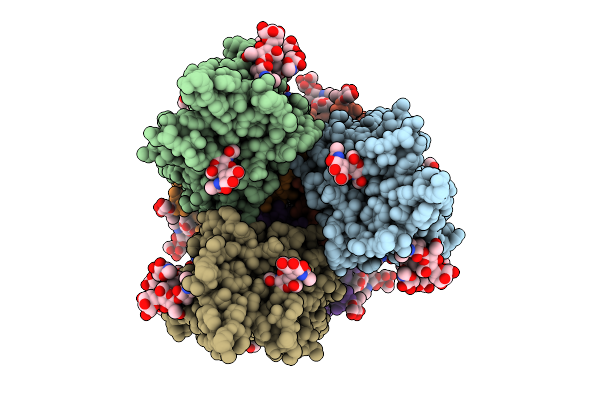
Pre-Fusion Herv-K Envelope Protein Trimer Ectodomain In Complex With Kenv-6 Fab
Organism: Homo sapiens, Enterobacteria phage t4, Mus musculus
Method: ELECTRON MICROSCOPY Release Date: 2025-07-30 Classification: VIRAL PROTEIN Ligands: NAG |
 |
Organism: Homo sapiens, Enterobacteria phage t4
Method: ELECTRON MICROSCOPY Release Date: 2025-07-30 Classification: VIRAL PROTEIN Ligands: NAG |

