Search Count: 15
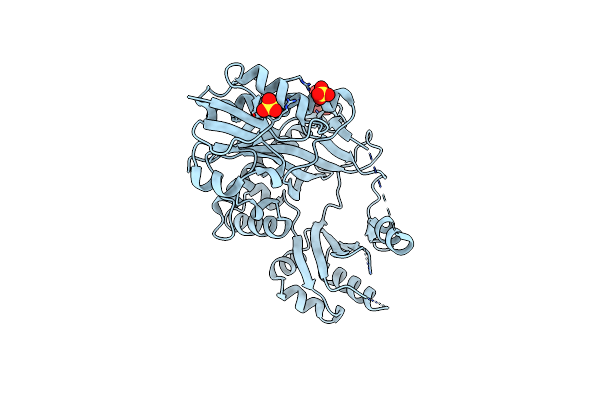 |
Crystal Structure Of The C-Terminal Domain From K. Lactis Pby1, An Atp-Grasp Enzyme Interacting With The Mrna Decapping Enzyme Dcp2
Organism: Kluyveromyces lactis (strain atcc 8585 / cbs 2359 / dsm 70799 / nbrc 1267 / nrrl y-1140 / wm37)
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2020-04-29 Classification: LIGASE Ligands: SO4 |
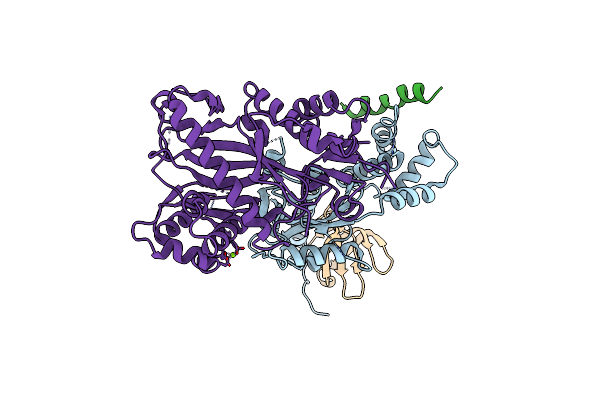 |
Crystal Structure Of The Pby1 Atp-Grasp Enzyme Bound To The S. Cerevisiae Mrna Decapping Complex (Dcp1-Dcp2-Edc3)
Organism: Saccharomyces cerevisiae s288c
Method: X-RAY DIFFRACTION Resolution:3.49 Å Release Date: 2020-04-29 Classification: HYDROLASE Ligands: MG |
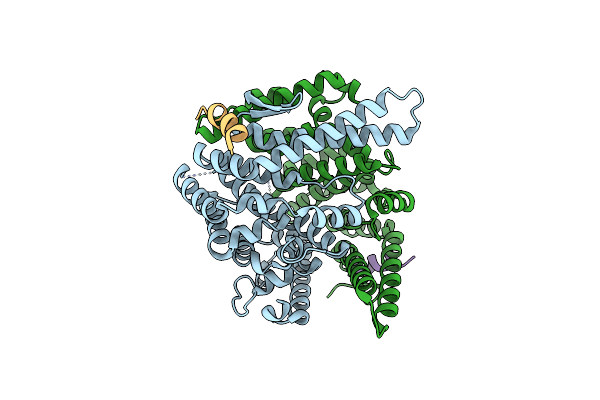 |
Structure Of C-Terminal Domain From S. Cerevisiae Pat1 Decapping Activator Bound To Dcp2 Hlm2 Peptide (Region 435-451)
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2017-08-16 Classification: RNA BINDING PROTEIN |
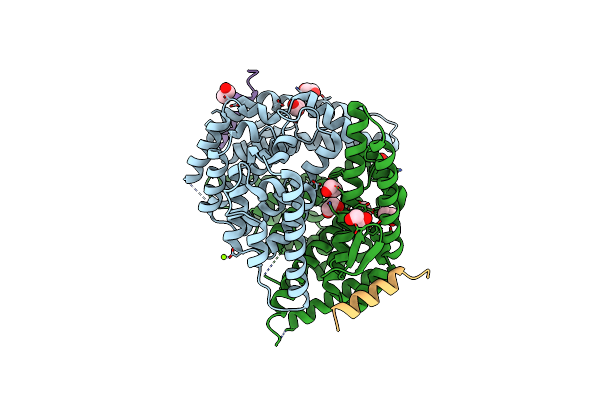 |
Structure Of C-Terminal Domain From S. Cerevisiae Pat1 Decapping Activator Bound To Dcp2 Hlm3 Peptide (Region 484-500)
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:2.15 Å Release Date: 2017-08-16 Classification: RNA BINDING PROTEIN Ligands: MG, CL, EDO |
 |
Structure Of C-Terminal Domain From S. Cerevisiae Pat1 Decapping Activator Bound To Dcp2 Hlm10 Peptide (Region 954-970)
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:1.89 Å Release Date: 2017-08-16 Classification: RNA BINDING PROTEIN Ligands: EDO, MG, TRS |
 |
Organism: Kluyveromyces lactis nrrl y-1140
Method: X-RAY DIFFRACTION Resolution:3.50 Å Release Date: 2016-10-05 Classification: RNA BINDING PROTEIN |
 |
Structure Of The Active Form Of /K. Lactis/ Dcp1-Dcp2-Edc3 Decapping Complex Bound To M7Gdp
Organism: Kluyveromyces lactis nrrl y-1140
Method: X-RAY DIFFRACTION Resolution:3.50 Å Release Date: 2016-10-05 Classification: RNA BINDING PROTEIN Ligands: MG, M7G |
 |
Nmr Solution Structure Of The Yeast Pih1 And Tah1 C-Terminal Domains Complex
Organism: Saccharomyces cerevisiae
Method: SOLUTION NMR Release Date: 2015-08-05 Classification: PROTEIN BINDING |
 |
Structure Of C-Terminal Domain From S. Cerevisiae Pat1 Decapping Activator (Space Group : P21)
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:2.15 Å Release Date: 2014-10-08 Classification: RNA BINDING PROTEIN Ligands: MES, EDO |
 |
Structure Of C-Terminal Domain From S. Cerevisiae Pat1 Decapping Activator (Space Group : P212121)
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:2.32 Å Release Date: 2014-10-08 Classification: CELL CYCLE Ligands: MG, EDO, CL |
 |
Organism: Saccharomyces cerevisiae
Method: SOLUTION NMR Release Date: 2014-09-10 Classification: PROTEIN BINDING |
 |
Organism: Saccharomyces cerevisiae
Method: SOLUTION NMR Release Date: 2013-11-27 Classification: RNA BINDING PROTEIN |
 |
Organism: Saccharomyces cerevisiae
Method: SOLUTION NMR Release Date: 2013-05-22 Classification: PROTEIN BINDING |
 |
The Nmr High Resolution Structure Of Yeast Tah1 In Complex With The Hsp90 C-Terminal Tail
Organism: Saccharomyces cerevisiae
Method: SOLUTION NMR Release Date: 2013-05-22 Classification: CHAPERONE-BINDING PROTEIN/CHAPERONE |
 |
The Structure Of 6-Phosphogluconate Dehydrogenase Refined At 2 Angstroms Resolution
Organism: Ovis aries
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 1995-02-27 Classification: OXIDOREDUCTASE (CHOH(D)-NADP+(A)) Ligands: SO4 |

