Search Count: 1,657
All
Selected
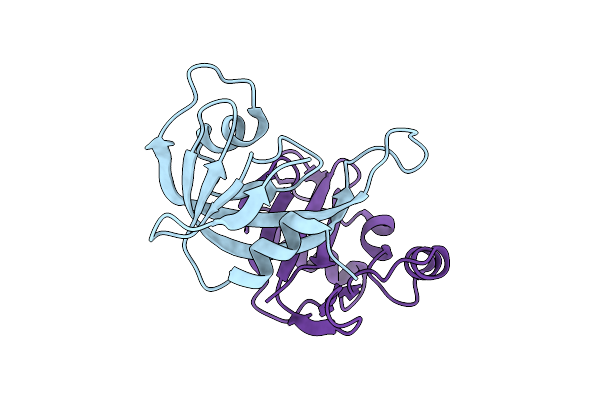 |
Crystal Structure For Yxid-Yxxd Toxin-Immunity Protein Complex From Bacillus Subtilis 6633.
Organism: Bacillus spizizenii atcc 6633 = jcm 2499
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2025-12-24 Classification: TOXIN |
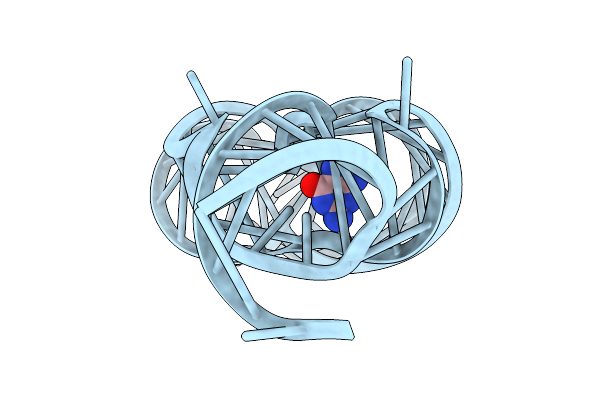 |
Organism: Paenibacillus sp. fsl e2-0178
Method: X-RAY DIFFRACTION Resolution:2.75 Å Release Date: 2025-11-26 Classification: RNA Ligands: GUN, MG |
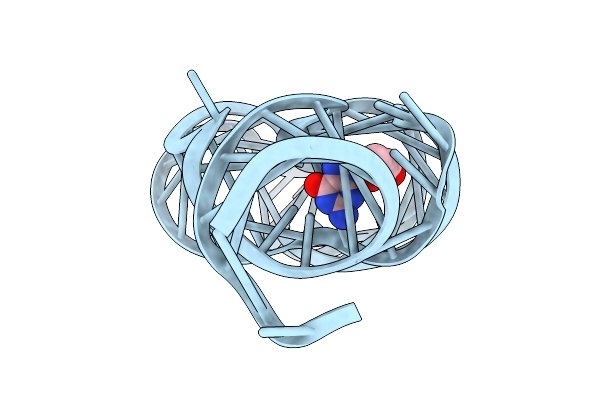 |
Crystal Structure Of Guanine-Ii Riboswitch In Complex With 2'-Deoxyguanosine
Organism: Paenibacillus sp. fsl e2-0178
Method: X-RAY DIFFRACTION Resolution:2.61 Å Release Date: 2025-11-26 Classification: RNA Ligands: MG, GNG |
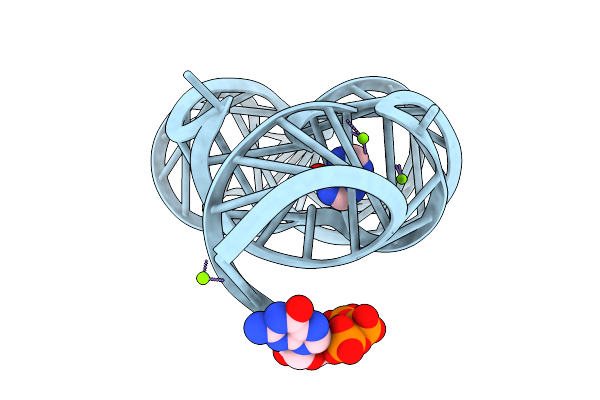 |
Organism: Paenibacillus sp. fsl e2-0178
Method: X-RAY DIFFRACTION Resolution:2.26 Å Release Date: 2025-11-26 Classification: RNA Ligands: HPA, GTP, MG |
 |
Organism: Paenibacillus sp. fsl e2-0178
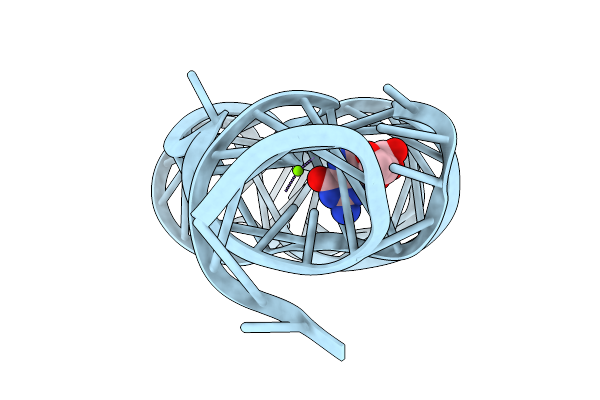
Method: X-RAY DIFFRACTION Resolution:2.83 Å Release Date: 2025-11-26 Classification: RNA Ligands: GMP, MG |
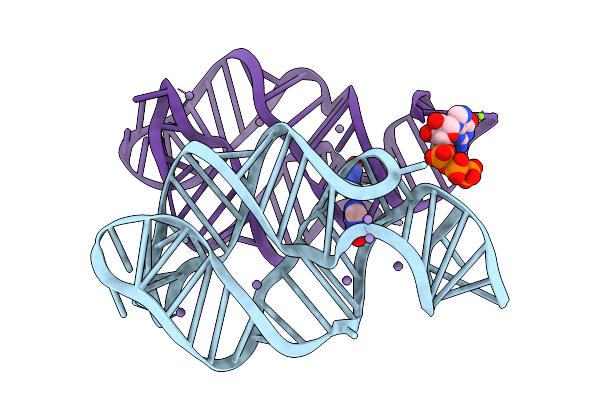 |
Crystal Structure Of Guanine-Ii Riboswitch In Complex With Guanine Soaked With Mn2+
Organism: Paenibacillus sp. fsl e2-0178
Method: X-RAY DIFFRACTION Resolution:2.97 Å Release Date: 2025-11-26 Classification: RNA Ligands: GUN, MN, GTP, MG |
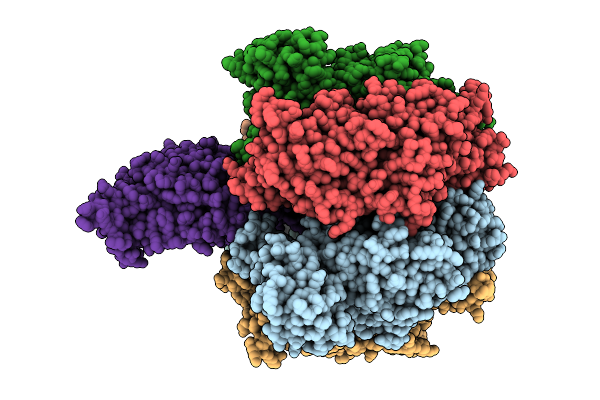 |
Organism: Bacillus sp. ps3
Method: ELECTRON MICROSCOPY Release Date: 2025-11-05 Classification: ELECTRON TRANSPORT Ligands: MG, ATP, PO4, ADP |
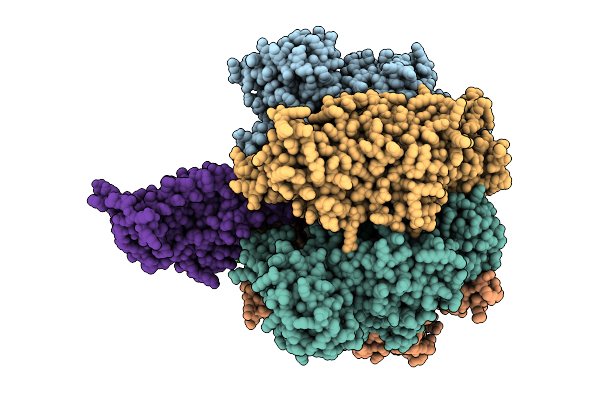 |
Organism: Bacillus sp. ps3, Pseudomonas aeruginosa
Method: ELECTRON MICROSCOPY Release Date: 2025-11-05 Classification: ELECTRON TRANSPORT Ligands: ATP, MG, ADP |
 |
Monomeric Sarcosine Oxidase From Bacillus Sp. (Soxb) Complexed With L-Thioproline
Organism: Bacillus sp. b-0618
Method: X-RAY DIFFRACTION Resolution:1.30 Å Release Date: 2025-09-24 Classification: OXIDOREDUCTASE Ligands: FAD, PRS, CL, GOL, SO4 |
 |
Monomeric Sarcosine Oxidase From Bacillus Sp. (Soxb) Complexed With L-Proline
Organism: Bacillus sp. b-0618
Method: X-RAY DIFFRACTION Resolution:1.54 Å Release Date: 2025-09-24 Classification: OXIDOREDUCTASE Ligands: FAD, PRO, GOL, CL, SO4, PO4 |
 |
Monomeric Sarcosine Oxidase From Bacillus Sp. (Soxb) Complexed With D-Proline
Organism: Bacillus sp. b-0618
Method: X-RAY DIFFRACTION Resolution:1.35 Å Release Date: 2025-09-24 Classification: OXIDOREDUCTASE Ligands: FAD, DPR, CL, GOL, SO4 |
 |
Organism: Bacillus sp. b-0618
Method: X-RAY DIFFRACTION Resolution:1.35 Å Release Date: 2025-09-24 Classification: OXIDOREDUCTASE Ligands: FAD, GOL, CL, NA |
 |
Organism: Bacillus sp. b-0618
Method: X-RAY DIFFRACTION Resolution:1.45 Å Release Date: 2025-09-24 Classification: OXIDOREDUCTASE Ligands: FAD, GOL, CL |
 |
Organism: Bacillus sp. ps3
Method: ELECTRON MICROSCOPY Release Date: 2025-07-09 Classification: MEMBRANE PROTEIN Ligands: ADP, MG |
 |
Organism: Bacillus sp. ps3
Method: ELECTRON MICROSCOPY Release Date: 2025-07-09 Classification: MEMBRANE PROTEIN |
 |
Organism: Bacillus sp. (in: firmicutes)
Method: X-RAY DIFFRACTION Resolution:1.49 Å Release Date: 2025-06-18 Classification: HYDROLASE Ligands: GOL, MG, ZN |
 |
Organism: Bacillus sp. hmf5848
Method: ELECTRON MICROSCOPY Release Date: 2025-06-04 Classification: ANTIVIRAL PROTEIN |
 |
Organism: Bacillus sp. hmf5848
Method: ELECTRON MICROSCOPY Release Date: 2025-06-04 Classification: ANTIVIRAL PROTEIN |
 |
Organism: Bacillus sp. hmf5848
Method: ELECTRON MICROSCOPY Release Date: 2025-06-04 Classification: ANTIVIRAL PROTEIN |
 |
Organism: Bacillus sp. hmf5848
Method: ELECTRON MICROSCOPY Release Date: 2025-06-04 Classification: ANTIVIRAL PROTEIN |

