Search Count: 509
All
Selected
 |
Cyanide Dihydratase From Bacillus Pumilus C1 E155R Variant With Altered Helical Twist.
Organism: Bacillus pumilus
Method: ELECTRON MICROSCOPY Release Date: 2024-04-10 Classification: HYDROLASE |
 |
Organism: Bacillus pumilus
Method: ELECTRON MICROSCOPY Release Date: 2023-08-16 Classification: HYDROLASE |
 |
Organism: Bacillus pumilus
Method: X-RAY DIFFRACTION Resolution:1.10 Å Release Date: 2023-02-22 Classification: ISOMERASE Ligands: EDO |
 |
Cyanide Dihydratase From Bacillus Pumilus C1 Variant - Q86R,H305K,H308K,H323K
Organism: Bacillus pumilus
Method: ELECTRON MICROSCOPY Release Date: 2023-01-18 Classification: HYDROLASE |
 |
Organism: Bacillus pumilus
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2022-05-25 Classification: HYDROLASE Ligands: OAA, DEP |
 |
Organism: Bacillus pumilus
Method: X-RAY DIFFRACTION Resolution:0.87 Å Release Date: 2022-05-25 Classification: HYDROLASE Ligands: CIT, PO4, PPI |
 |
Organism: Bacillus pumilus safr-032
Method: X-RAY DIFFRACTION Resolution:2.99 Å Release Date: 2022-05-11 Classification: OXIDOREDUCTASE Ligands: CU |
 |
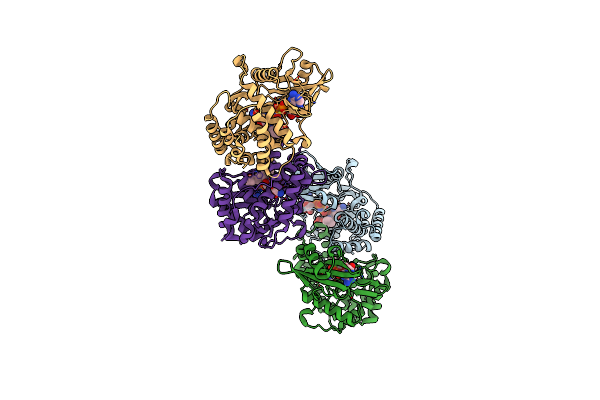
Organism: Bacillus pumilus
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2020-07-22 Classification: ANTIMICROBIAL PROTEIN Ligands: 1PE |
 |
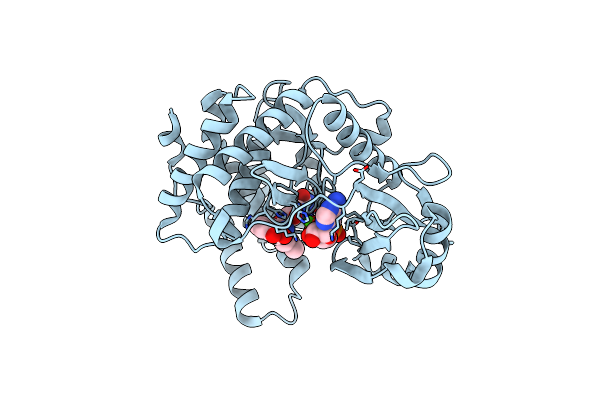
Organism: Bacillus pumilus
Method: X-RAY DIFFRACTION Resolution:1.35 Å Release Date: 2020-07-22 Classification: ANTIMICROBIAL PROTEIN Ligands: MG, CL, UAM, ANP |
 |
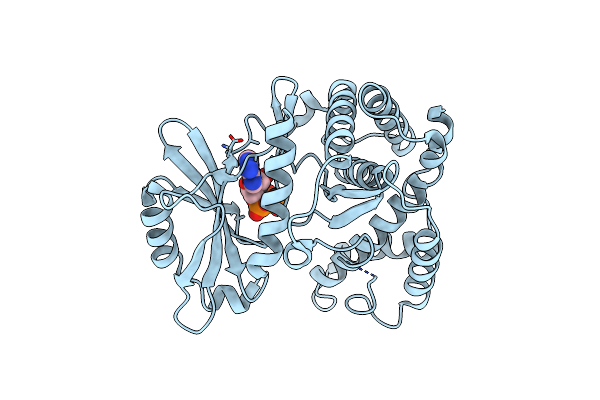
Organism: Bacillus pumilus
Method: X-RAY DIFFRACTION Resolution:1.35 Å Release Date: 2020-07-22 Classification: ANTIMICROBIAL PROTEIN Ligands: CA, ANP, UAM |
 |
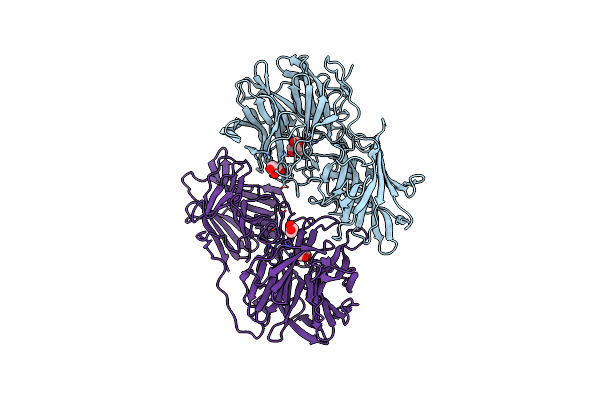
Organism: Bacillus pumilus
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2020-07-22 Classification: ANTIMICROBIAL PROTEIN Ligands: ATP |
 |
Organism: Bacillus pumilus
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2019-03-13 Classification: HYDROLASE Ligands: GOL |
 |
Structure Of B. Pumilus Gh48 In Complex With A Cellobio-Derived Isofagomine
Organism: Bacillus pumilus
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2018-10-31 Classification: HYDROLASE Ligands: G2I, 9MR, CA, NA, EDO, MLI, ACT |
 |
Organism: Bacillus pumilus
Method: X-RAY DIFFRACTION Resolution:1.73 Å Release Date: 2018-05-30 Classification: HYDROLASE Ligands: GOL |
 |
Crystal Structure Of Beta-Xylosidase Mutant (E186Q/F503Y) From Bacillus Pumilus
Organism: Bacillus pumilus
Method: X-RAY DIFFRACTION Resolution:1.78 Å Release Date: 2018-05-30 Classification: HYDROLASE Ligands: XYP |
 |
Organism: Bacillus pumilus
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2018-05-30 Classification: HYDROLASE |
 |
Crystal Structure Of Pyrene- And Phenanthrene-Modified Dna In Complex With The Bpuj1 Endonuclease Binding Domain
Organism: Bacillus pumilus, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.67 Å Release Date: 2016-08-17 Classification: HYDROLASE |
 |
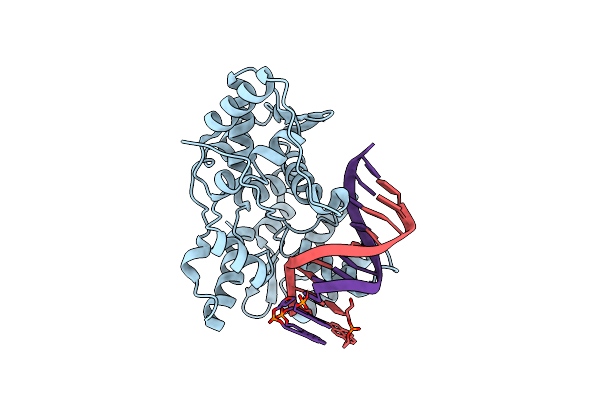
Crystal Structure Of Pyrene- And Phenanthrene-Modified Dna In Complex With The Bpuj1 Endonuclease Binding Domain
Organism: Bacillus pumilus, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.55 Å Release Date: 2016-08-17 Classification: HYDROLASE |
 |
Crystal Structure Of Pyrene- And Phenanthrene-Modified Dna In Complex With The Bpuj1 Endonuclease Binding Domain
Organism: Bacillus pumilus, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.88 Å Release Date: 2016-08-17 Classification: HYDROLASE |
 |
The Structure Of Bacillus Pumilus Gh48 In Complex With Cellobiose And Cellohexaose
Organism: Bacillus pumilus (strain safr-032)
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2016-08-10 Classification: HYDROLASE Ligands: CA, MLI, EDO, GOL, NA |

