Search Count: 6,735
All
Selected
 |
Organism: Bacillus subtilis subsp. subtilis str. 168
Method: X-RAY DIFFRACTION Resolution:2.64 Å Release Date: 2026-02-25 Classification: PROTEIN BINDING Ligands: ADP, AMP |
 |
Organism: Bacillus subtilis subsp. subtilis str. 168
Method: X-RAY DIFFRACTION Resolution:2.76 Å Release Date: 2026-02-25 Classification: DNA BINDING PROTEIN Ligands: ADP, PEG, CL |
 |
Organism: Bacillus subtilis subsp. subtilis str. 168
Method: X-RAY DIFFRACTION Resolution:3.35 Å Release Date: 2026-02-25 Classification: DNA BINDING PROTEIN Ligands: B4P, CL |
 |
Organism: Bacillus subtilis subsp. subtilis str. 168
Method: X-RAY DIFFRACTION Resolution:2.62 Å Release Date: 2026-02-25 Classification: DNA BINDING PROTEIN Ligands: ADP, PEG, CL, AMP |
 |
Organism: Bacillus subtilis subsp. subtilis str. 168
Method: X-RAY DIFFRACTION Resolution:3.06 Å Release Date: 2026-02-25 Classification: DNA BINDING PROTEIN Ligands: ADP, EDO, CL, PEG |
 |
Organism: Bacillus subtilis subsp. subtilis str. 168
Method: X-RAY DIFFRACTION Resolution:2.53 Å Release Date: 2026-02-25 Classification: DNA BINDING PROTEIN Ligands: ADP, ATP, EDO, CL, PEG |
 |
Organism: Bacillus subtilis subsp. subtilis str. 168
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2026-02-25 Classification: DNA BINDING PROTEIN Ligands: AMP, PEG, CL |
 |
Organism: Bacillus subtilis subsp. subtilis str. 168
Method: X-RAY DIFFRACTION Resolution:2.41 Å Release Date: 2026-02-18 Classification: BIOSYNTHETIC PROTEIN Ligands: B4P |
 |
Organism: Bacillus subtilis
Method: ELECTRON MICROSCOPY Release Date: 2026-01-21 Classification: DNA BINDING PROTEIN Ligands: ZN |
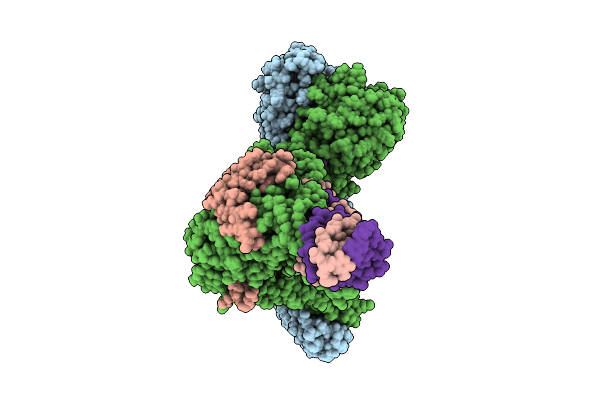 |
Attp Bound Large Serine Integrase And Rdf Complex In The Dimeric State (Cleaved)
Organism: Bacillus subtilis
Method: ELECTRON MICROSCOPY Release Date: 2026-01-21 Classification: DNA BINDING PROTEIN Ligands: ZN |
 |
Organism: Bacillus subtilis
Method: ELECTRON MICROSCOPY Release Date: 2026-01-21 Classification: DNA BINDING PROTEIN Ligands: ZN |
 |
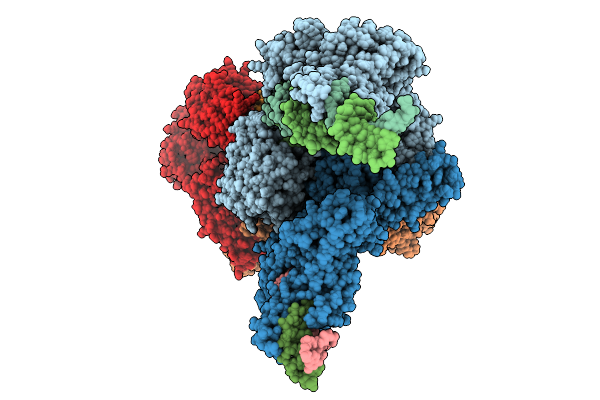
Attpmm And Attbmm Bound Serine Integrase Complex In The Post-Rotation State
Organism: Bacillus subtilis
Method: ELECTRON MICROSCOPY Release Date: 2026-01-21 Classification: DNA BINDING PROTEIN Ligands: ZN |
 |
Organism: Bacillus subtilis
Method: ELECTRON MICROSCOPY Release Date: 2026-01-21 Classification: DNA BINDING PROTEIN Ligands: ZN |
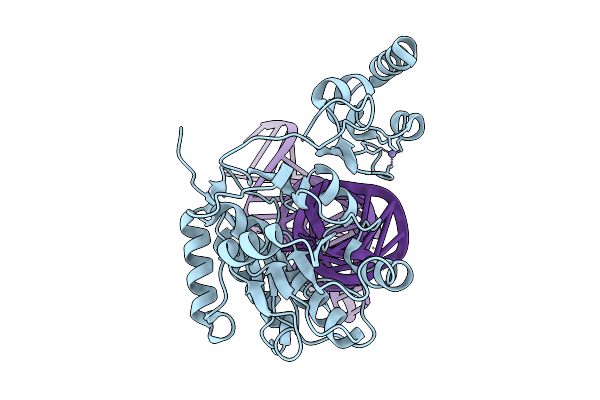 |
Organism: Bacillus subtilis subsp. subtilis str. 168
Method: ELECTRON MICROSCOPY Release Date: 2026-01-21 Classification: RNA BINDING PROTEIN Ligands: ZN |
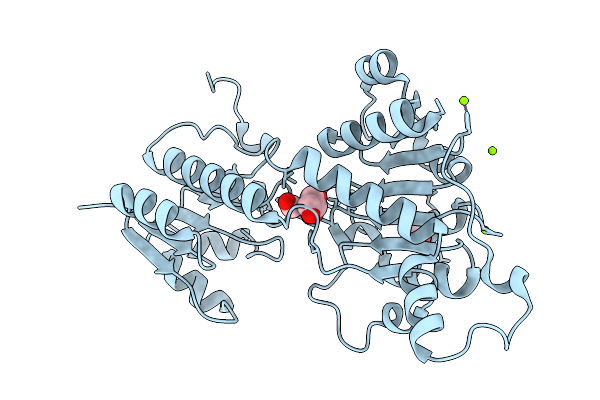 |
Crystal Structure Of Glyoxylate/Hydroxypyruvate Reductase In Complex From Bacillus Subtilis With Formate At Near-Atomic Resolution
Organism: Bacillus subtilis subsp. subtilis str. 168
Method: X-RAY DIFFRACTION Resolution:1.20 Å Release Date: 2026-01-21 Classification: OXIDOREDUCTASE Ligands: MG, FMT, GOL |
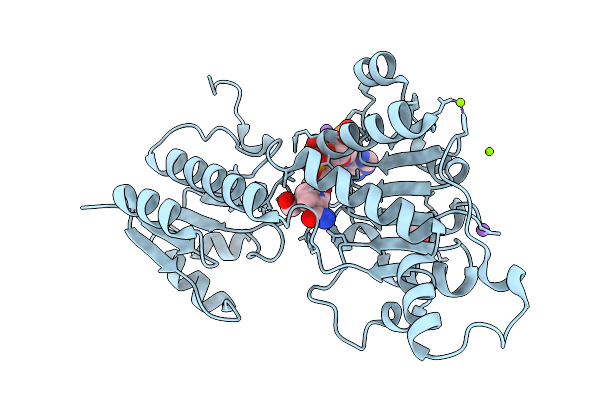 |
Crystal Structure Of Glyoxylate/Hydroxypyruvate Reductase In Complex From Bacillus Subtilis With Formate And Nadph At Near-Atomic Resolution
Organism: Bacillus subtilis subsp. subtilis str. 168
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2026-01-21 Classification: OXIDOREDUCTASE Ligands: MG, FMT, NAP, NA |
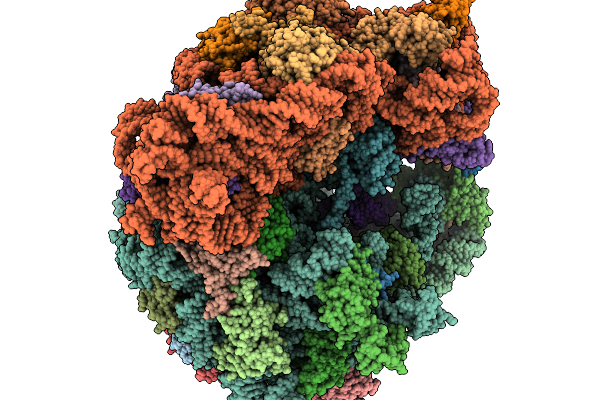 |
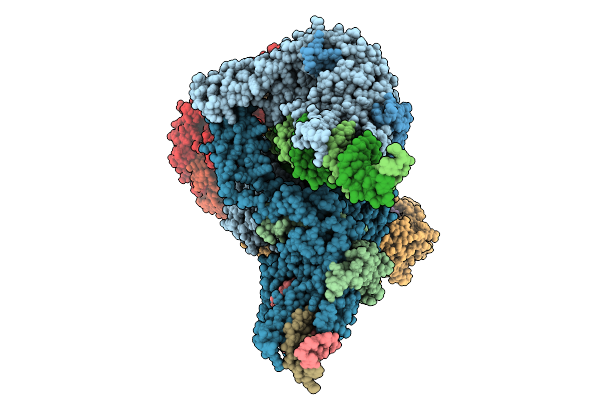
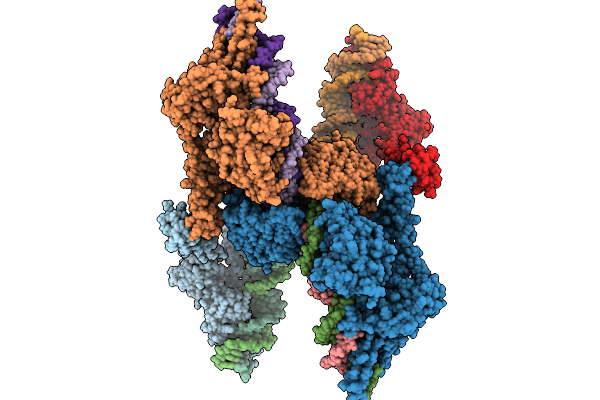
Structure Of Clim-Stalled Bacillus Subtilis 70S Ribosome With Release Factor Bound In The A-Site
Organism: Clostridioides difficile 630, Bacillus subtilis subsp. subtilis str. 168
Method: ELECTRON MICROSCOPY Resolution:2.30 Å Release Date: 2026-01-14 Classification: RIBOSOME Ligands: ZN |
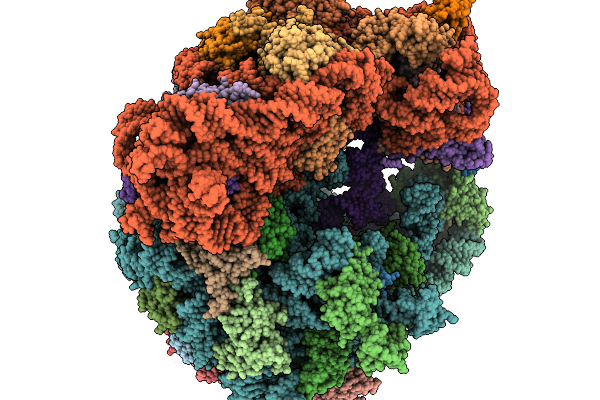 |
Organism: Clostridioides difficile 630, Bacillus subtilis subsp. subtilis str. 168
Method: ELECTRON MICROSCOPY Resolution:2.30 Å Release Date: 2026-01-14 Classification: RIBOSOME |
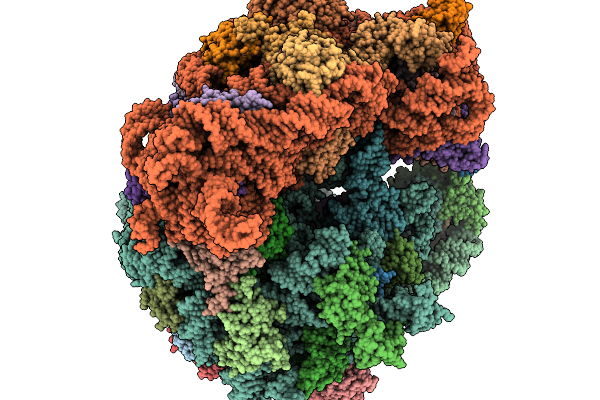 |
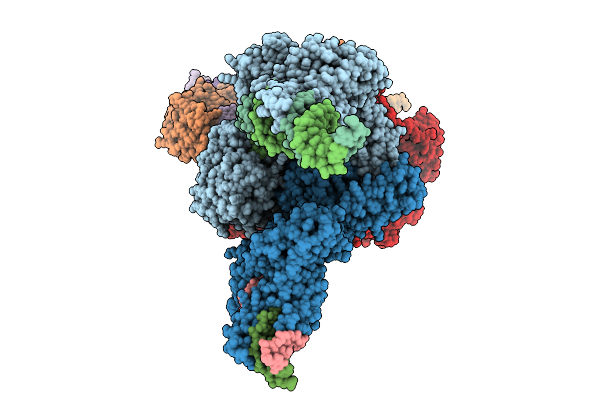
Structure Of Clim-Stalled Bacillus Subtilis 70S Ribosome With Trna-Tyr In The A-Site
Organism: Clostridioides difficile 630, Bacillus subtilis subsp. subtilis str. 168
Method: ELECTRON MICROSCOPY Resolution:2.80 Å Release Date: 2026-01-14 Classification: RIBOSOME Ligands: ZN |
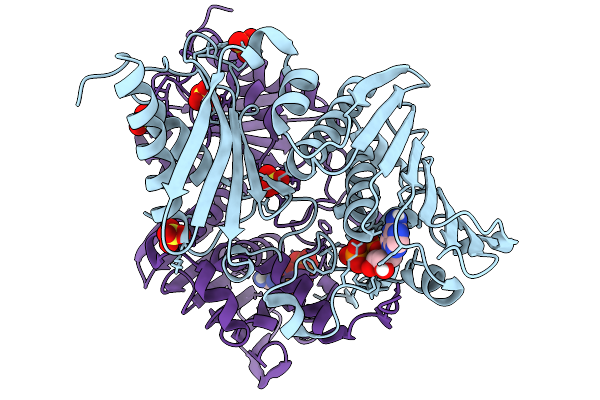 |
Organism: Bacillus subtilis
Method: X-RAY DIFFRACTION Resolution:1.36 Å Release Date: 2026-01-07 Classification: DNA BINDING PROTEIN Ligands: ADP, MG, SO4 |

