Search Count: 47
 |
Organism: Physeter catodon
Method: X-RAY DIFFRACTION Resolution:1.40 Å Release Date: 2023-12-06 Classification: OXYGEN STORAGE Ligands: SO4, WRK |
 |
Organism: Physeter catodon
Method: X-RAY DIFFRACTION Resolution:1.04 Å Release Date: 2023-12-06 Classification: OXYGEN STORAGE Ligands: HEM, IMD, SO4 |
 |
Organism: Physeter catodon
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2022-06-01 Classification: OXYGEN STORAGE Ligands: HEM, SO4 |
 |
Organism: Physeter catodon
Method: X-RAY DIFFRACTION Resolution:1.17 Å Release Date: 2022-06-01 Classification: OXYGEN STORAGE Ligands: NO3, HEM, OXY, SO4 |
 |
Crystal Structure Of Sperm Whale Myoglobin Variant Smb13(Pcaaf) In Space Group P212121
Organism: Physeter catodon
Method: X-RAY DIFFRACTION Resolution:1.30 Å Release Date: 2022-06-01 Classification: OXYGEN STORAGE Ligands: HEM, SO4, CL |
 |
Crystal Structure Of Sperm Whale Myoglobin Variant Smb13(Pcaaf) In Space Group P21
Organism: Physeter macrocephalus
Method: X-RAY DIFFRACTION Resolution:1.30 Å Release Date: 2022-06-01 Classification: OXYGEN STORAGE Ligands: HEM, OXY, SO4 |
 |
Dark-Operative Protochlorophyllide Oxidoreductase In The Nucleotide-Free Form.
Organism: Rhodobacter sphaeroides
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2020-12-02 Classification: OXIDOREDUCTASE Ligands: SF4, CL |
 |
X-Ray Crystal Structure Of Bacillus Subtilis Ribonucleotide Reductase Nrde Alpha Subunit With Ttp, Atp, And Adp
Organism: Bacillus subtilis
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2019-06-19 Classification: OXIDOREDUCTASE Ligands: TTP, ADP, ATP, MG |
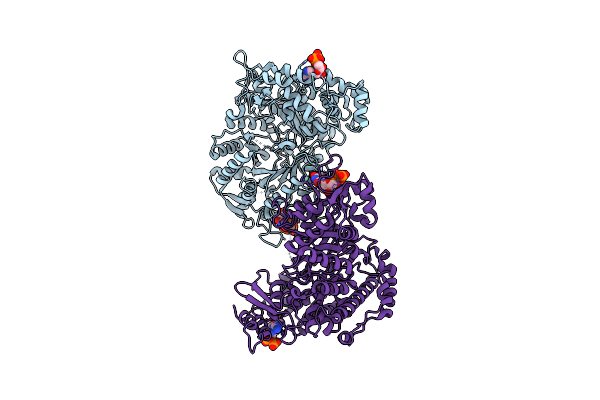 |
X-Ray Crystal Structure Of Bacillus Subtilis Ribonucleotide Reductase Nrde Alpha Subunit With Ttp And Adp
Organism: Bacillus subtilis
Method: X-RAY DIFFRACTION Resolution:2.95 Å Release Date: 2019-06-19 Classification: OXIDOREDUCTASE Ligands: TTP, ADP, MG |
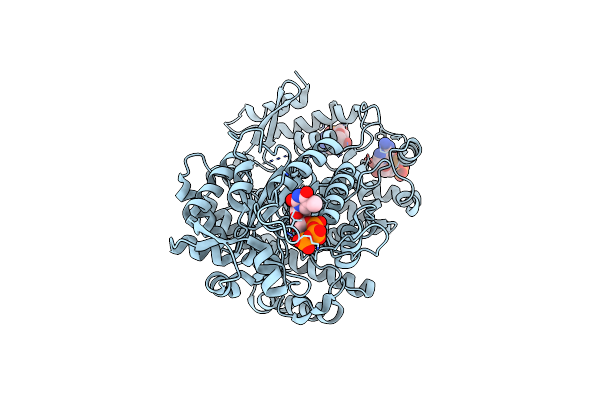 |
Reduced X-Ray Crystal Structure Of Bacillus Subtilis Ribonucleotide Reductase Nrde Alpha Subunit With Ttp, Atp, And Adp
Organism: Bacillus subtilis
Method: X-RAY DIFFRACTION Resolution:2.55 Å Release Date: 2019-06-19 Classification: OXIDOREDUCTASE Ligands: TTP, ADP, ATP, MG |
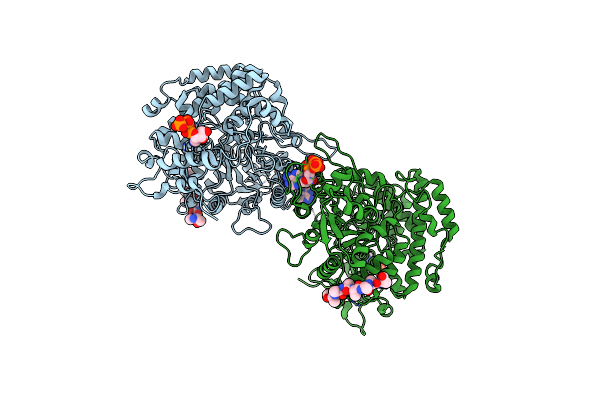 |
Em Structure Of Bacillus Subtilis Ribonucleotide Reductase Inhibited Filament Composed Of Nrde Alpha Subunit And Nrdf Beta Subunit With Datp
Organism: Bacillus subtilis
Method: ELECTRON MICROSCOPY Release Date: 2019-06-19 Classification: OXIDOREDUCTASE Ligands: DTP |
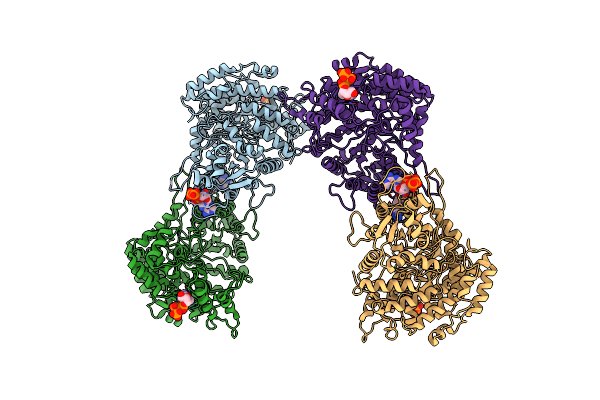 |
Em Structure Of Bacillus Subtilis Ribonucleotide Reductase Inhibited Double-Helical Filament Of Nrde Alpha Subunit With Datp
Organism: Bacillus subtilis
Method: ELECTRON MICROSCOPY Release Date: 2019-06-19 Classification: OXIDOREDUCTASE, PROTEIN FIBRIL Ligands: DTP |
 |
Organism: Physeter catodon
Method: X-RAY DIFFRACTION Resolution:1.10 Å Release Date: 2019-01-23 Classification: TRANSFERASE Ligands: SO4, HEM |
 |
Organism: Streptococcus suis
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2017-08-30 Classification: METAL BINDING PROTEIN Ligands: SF4 |
 |
Organism: Streptococcus suis
Method: X-RAY DIFFRACTION Resolution:2.49 Å Release Date: 2017-08-30 Classification: METAL BINDING PROTEIN Ligands: SF4, SAM |
 |
Crystal Structure Of Streptococcus Suis Suib Bound To Precursor Peptide Suia
Organism: Streptococcus suis
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2017-08-30 Classification: METAL BINDING PROTEIN Ligands: SF4, MET, SAM |
 |
Room Temperature X-Ray Crystallographic Structure Of A Jonesia Denitrificans Lytic Polysaccharide Monooxygenase At 1.1 Angstrom Resolution.
Organism: Jonesia denitrificans (strain atcc 14870 / dsm 20603 / cip 55134)
Method: X-RAY DIFFRACTION Resolution:1.10 Å Release Date: 2017-05-24 Classification: SUGAR BINDING PROTEIN Ligands: CU, PER |
 |
Neutron Crystallographic Structure Of A Jonesia Denitrificans Lytic Polysaccharide Monooxygenase
Organism: Jonesia denitrificans (strain atcc 14870 / dsm 20603 / cip 55134)
Method: NEUTRON DIFFRACTION Resolution:2.10 Å Release Date: 2017-05-24 Classification: SUGAR BINDING PROTEIN Ligands: CU, PER |
 |
Organism: Lipomyces starkeyi
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2017-02-08 Classification: TRANSFERASE Ligands: ADP, MG, SO4, K |
 |
Crystal Structure Of A Highly Thermal Stable But Inactive Levoglucosan Kinase.
Organism: Lipomyces starkeyi
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2015-09-23 Classification: DE NOVO PROTEIN |

