Search Count: 17
 |
Mosquitocidal Cry11Aa Determined At Ph 7 From Naturally-Occurring Nanocrystals By Serial Femtosecond Crystallography
Organism: Bacillus thuringiensis serovar israelensis
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2022-07-27 Classification: TOXIN |
 |
Mosquitocidal Cry11Aa-Y449F Determined At Ph 7 From Naturally-Occurring Nanocrystals By Serial Femtosecond Crystallography
Organism: Bacillus thuringiensis serovar israelensis
Method: X-RAY DIFFRACTION Resolution:3.10 Å Release Date: 2022-07-27 Classification: TOXIN |
 |
Mosquitocidal Cry11Aa-E583Q Determined At Ph 7 From Naturally-Occurring Nanocrystals By Serial Femtosecond Crystallography
Organism: Bacillus thuringiensis serovar israelensis
Method: X-RAY DIFFRACTION Resolution:3.30 Å Release Date: 2022-07-27 Classification: TOXIN |
 |
Mosquitocidal Cry11Aa-F17Y Determined At Ph 7 From Naturally-Occurring Nanocrystals By Serial Femtosecond Crystallography
Organism: Bacillus thuringiensis serovar israelensis
Method: X-RAY DIFFRACTION Resolution:3.40 Å Release Date: 2022-07-27 Classification: TOXIN |
 |
Mosquitocidal Cry11Ba Determined At Ph 6.5 From Naturally-Occurring Nanocrystals By Serial Femtosecond Crystallography
Organism: Bacillus thuringiensis serovar jegathesan
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2022-07-27 Classification: TOXIN |
 |
Mosquitocidal Cry11Ba Determined At Ph 10.4 From Naturally-Occurring Nanocrystals By Serial Femtosecond Crystallography
Organism: Bacillus thuringiensis serovar jegathesan
Method: X-RAY DIFFRACTION Resolution:2.65 Å Release Date: 2022-07-27 Classification: TOXIN Ligands: GOL |
 |
Organism: Streptomyces rubiginosus
Method: X-RAY DIFFRACTION Resolution:2.13 Å Release Date: 2021-06-16 Classification: ISOMERASE Ligands: MN |
 |
Native Structure Of Mosquitocidal Cyt1A Protoxin Obtained By Serial Femtosecond Crystallography On In Vivo Grown Crystals At Ph 7
Organism: Bacillus thuringiensis subsp. israelensis
Method: X-RAY DIFFRACTION Resolution:1.86 Å Release Date: 2020-10-14 Classification: TOXIN |
 |
Structure Of Mosquitocidal Cyt1A Protoxin Obtained By Serial Femtosecond Crystallography On In Vivo Grown Crystals Soaked With Dtt At Ph 7
Organism: Bacillus thuringiensis subsp. israelensis
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2020-10-14 Classification: TOXIN |
 |
Structure Of Mosquitocidal Cyt1Aa Protoxin Obtained By Serial Femtosecond Crystallography On In Vivo Grown Crystals At Ph 10
Organism: Bacillus thuringiensis subsp. israelensis
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2020-10-14 Classification: TOXIN Ligands: CA |
 |
Structure Of The C7S Mutant Of Mosquitocidal Cyt1A Protoxin Obtained By Serial Femtosecond Crystallography On In Vivo Grown Crystals At Ph 7
Organism: Bacillus thuringiensis subsp. israelensis
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2020-10-14 Classification: TOXIN |
 |
Organism: Escherichia phage t5
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2018-01-03 Classification: VIRAL PROTEIN Ligands: CL |
 |
Organism: Escherichia coli k-12
Method: ELECTRON MICROSCOPY Resolution:6.10 Å Release Date: 2016-09-21 Classification: LYASE |
 |
Organism: Escherichia coli
Method: ELECTRON MICROSCOPY Resolution:5.50 Å Release Date: 2016-09-21 Classification: LYASE |
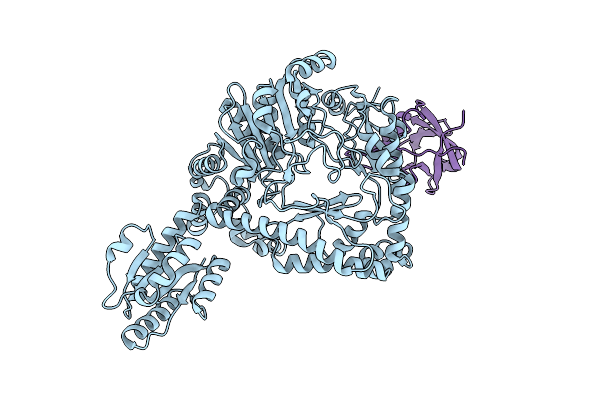 |
Revisited Cryo-Em Structure Of Inducible Lysine Decarboxylase Complexed With Lara Domain Of Rava Atpase
Organism: Escherichia coli k-12
Method: ELECTRON MICROSCOPY Resolution:6.20 Å Release Date: 2016-09-21 Classification: HYDROLASE/ISOMERASE |
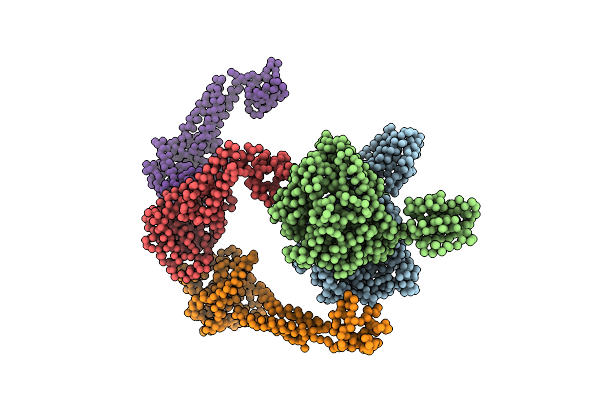 |
Electron Cryo-Microscopy Of The Complex Formed Between The Hexameric Atpase Rava And The Decameric Inducible Decarboxylase Ldci
Organism: Escherichia coli k-12
Method: ELECTRON MICROSCOPY Resolution:11.00 Å Release Date: 2014-08-20 Classification: LYASE/HYDROLASE |
 |
Assembly Principles Of The Unique Cage Formed By The Atpase Rava Hexamer And The Lysine Decarboxylase Ldci Decamer
Organism: Escherichia coli str. k-12 substr. mg1655
Method: ELECTRON MICROSCOPY Resolution:7.50 Å Release Date: 2014-08-20 Classification: LYASE/HYDROLASE |

