Search Count: 4
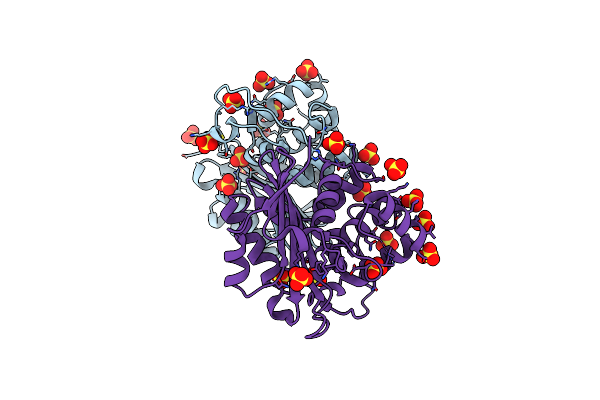 |
Crystal Structure Of The Class D Beta-Lactamase Oxa-935 From Pseudomonas Aeruginosa, Orthorhombic Crystal Form
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Resolution:1.96 Å Release Date: 2022-07-06 Classification: HYDROLASE Ligands: SO4 |
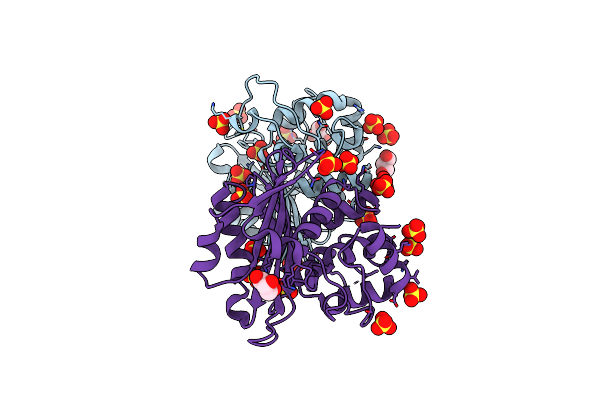 |
Crystal Structure Of The Oxacillin-Hydrolyzing Class D Extended-Spectrum Beta-Lactamase Oxa-14 From Pseudomonas Aeruginosa
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2021-12-29 Classification: HYDROLASE Ligands: GOL, SO4 |
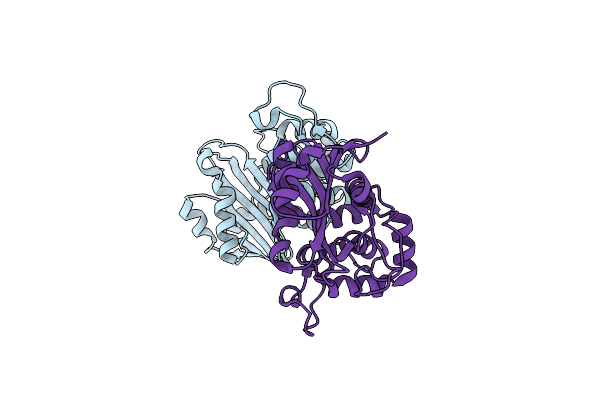 |
Crystal Structure Of The Class D Beta-Lactamase Oxa-935 From Pseudomonas Aeruginosa, Monoclinic Crystal Form
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Resolution:1.30 Å Release Date: 2021-12-29 Classification: HYDROLASE |
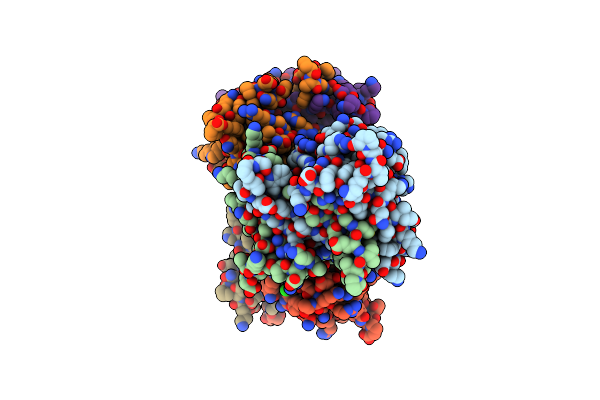 |
2.05 Angstrom Resolution Crystal Structure Of C-Terminal Dimerization Domain Of Nucleocapsid Phosphoprotein From Sars-Cov-2
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Resolution:2.05 Å Release Date: 2020-04-22 Classification: VIRAL PROTEIN Ligands: CL |

