Search Count: 44
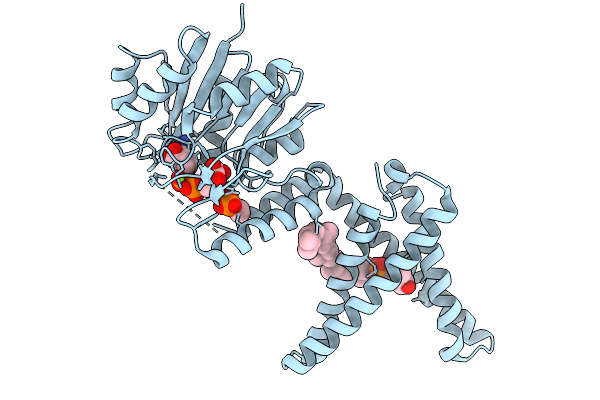 |
Dolichyl Phosphate Mannose Synthase In Complex With Donor (Gdp-Man) And Traces Of Acceptor (Dol55P) And Product (Dol55P-Man)
Organism: Pyrococcus furiosus dsm 3638
Method: X-RAY DIFFRACTION Release Date: 2025-10-01 Classification: TRANSFERASE Ligands: GDD, MG, EFS, MJC |
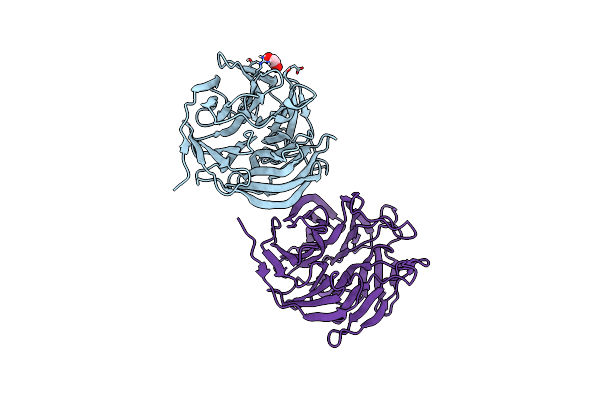 |
Organism: Loligo vulgaris
Method: X-RAY DIFFRACTION Release Date: 2025-05-14 Classification: BIOSYNTHETIC PROTEIN Ligands: EDO |
 |
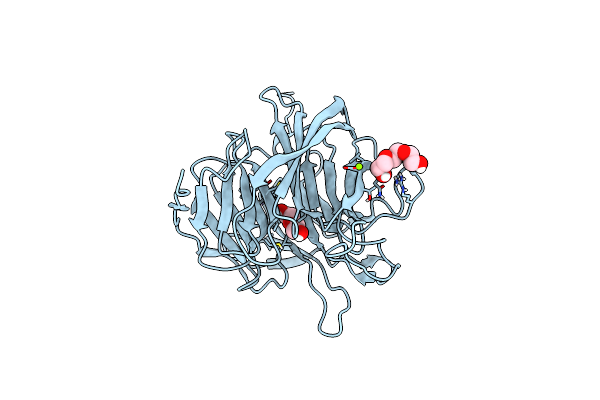
Organism: Loligo vulgaris
Method: X-RAY DIFFRACTION Release Date: 2025-05-14 Classification: BIOSYNTHETIC PROTEIN Ligands: PEG, EDO |
 |
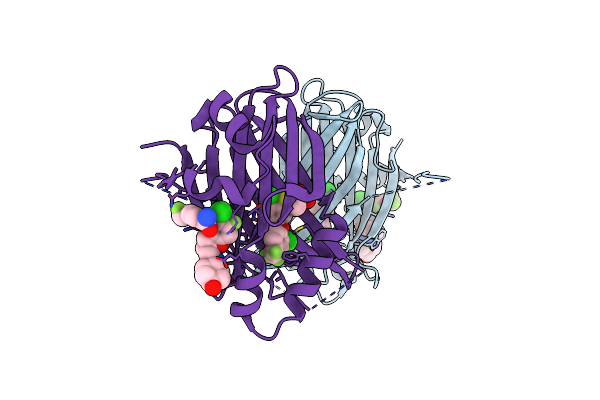
Organism: Loligo vulgaris
Method: X-RAY DIFFRACTION Release Date: 2025-05-14 Classification: BIOSYNTHETIC PROTEIN Ligands: PEG, PGE, MG |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2023-04-12 Classification: TRANSCRIPTION Ligands: U3F, U3O, PO4 |
 |
Organism: Mus musculus
Method: X-RAY DIFFRACTION Resolution:2.31 Å Release Date: 2022-12-21 Classification: HYDROLASE Ligands: ACT, SO4, GOL, CA |
 |
Organism: Mus musculus
Method: X-RAY DIFFRACTION Resolution:3.16 Å Release Date: 2022-12-21 Classification: HYDROLASE Ligands: 7VK, GOL, K |
 |
Wild-Type Ferulic Acid Esterase From Lactobacillus Buchneri In Complex With Ferulate
Organism: Lentilactobacillus buchneri
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2022-11-30 Classification: HYDROLASE Ligands: FER, CA |
 |
Organism: Lentilactobacillus buchneri
Method: X-RAY DIFFRACTION Resolution:1.45 Å Release Date: 2022-11-30 Classification: HYDROLASE Ligands: CA, ACT |
 |
Organism: Lentilactobacillus buchneri
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2022-11-30 Classification: HYDROLASE |
 |
Organism: Opitutaceae bacterium tav5
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2021-07-14 Classification: HYDROLASE |
 |
Crystal Structure Of Brassica Juncea Hmg-Coa Synthase 1 Mutant - S359A In Complex With Acetyl-Coa
Organism: Brassica juncea
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2021-07-14 Classification: TRANSFERASE Ligands: ACO, A3P |
 |
Exo-Beta-1,3-Glucanase From Moose Rumen Microbiome, Active Site Mutant E167Q/E295Q
Organism: Uncultured bacterium
Method: X-RAY DIFFRACTION Resolution:1.35 Å Release Date: 2021-01-13 Classification: HYDROLASE Ligands: 15P |
 |
Organism: Uncultured bacterium
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2021-01-13 Classification: HYDROLASE |
 |
Crystal Structure Of Puf60 Uhm Domain In Complex With 7,8 Dimethoxyperphenazine
Organism: Citrobacter freundii mgh 56, Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.94 Å Release Date: 2020-09-09 Classification: SPLICING Ligands: MG, LJT |
 |
Organism: Pyrobaculum calidifontis (strain jcm 11548 / va1)
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2020-07-22 Classification: TRANSFERASE |
 |
Mannosyltransferase Pcmangt From Pyrobaculum Calidifontis In Complex With Gdp And Mn2+
Organism: Pyrobaculum calidifontis (strain jcm 11548 / va1)
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2020-07-22 Classification: TRANSFERASE Ligands: GDP, MN |
 |
Mannosyltransferase Pcmangt From Pyrobaculum Calidifontis In Complex With Gdp-Man And Mn2+
Organism: Pyrobaculum calidifontis (strain jcm 11548 / va1)
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2020-07-22 Classification: TRANSFERASE Ligands: GDD, MN |
 |
Organism: Cutibacterium acnes
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2019-07-03 Classification: HYDROLASE Ligands: MPO |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.33 Å Release Date: 2017-11-08 Classification: OXIDOREDUCTASE Ligands: NDP, C81, FLC |

