Search Count: 172
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-12-17 Classification: CYTOKINE Ligands: A1JPQ, SO4 |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-12-17 Classification: CYTOKINE Ligands: A1JPS, SO4 |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-12-17 Classification: CYTOKINE Ligands: A1JPU |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-09-17 Classification: HYDROLASE |
 |
Organism: Trichoplax adhaerens
Method: ELECTRON MICROSCOPY Release Date: 2025-09-17 Classification: HYDROLASE |
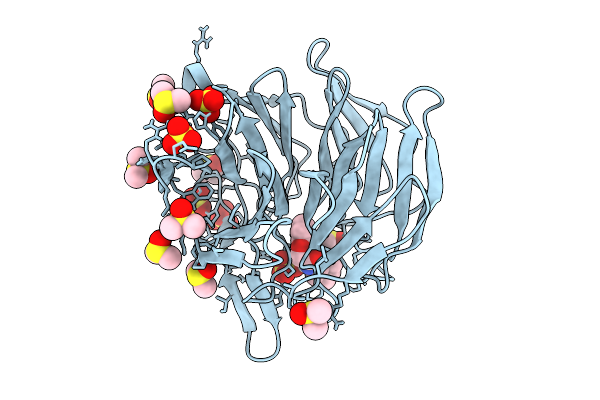 |
Crystal Structure Of The Keap1 Kelch Domain In Complex With The Xchem Fragment Z19735904 At 1.14 Angstrom Resolution.
Organism: Mus musculus
Method: X-RAY DIFFRACTION Release Date: 2025-09-03 Classification: PEPTIDE BINDING PROTEIN Ligands: B0A, SO4, DMS |
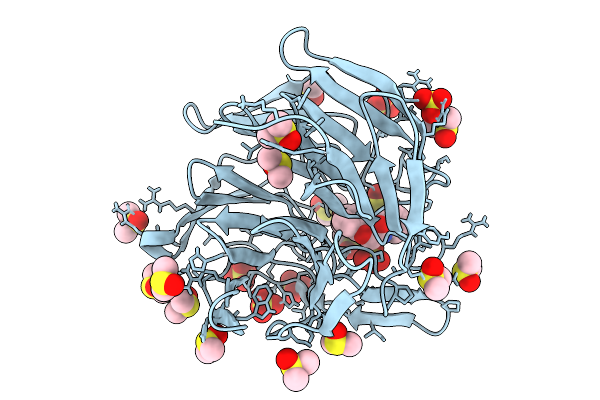 |
Crystal Structure Of The Keap1 Kelch Domain In Complex With The Small Molecule Ucab#827 At 1.40 Angstrom Resolution
Organism: Mus musculus
Method: X-RAY DIFFRACTION Release Date: 2025-09-03 Classification: PEPTIDE BINDING PROTEIN Ligands: A1IX2, CL, SO4, DMS |
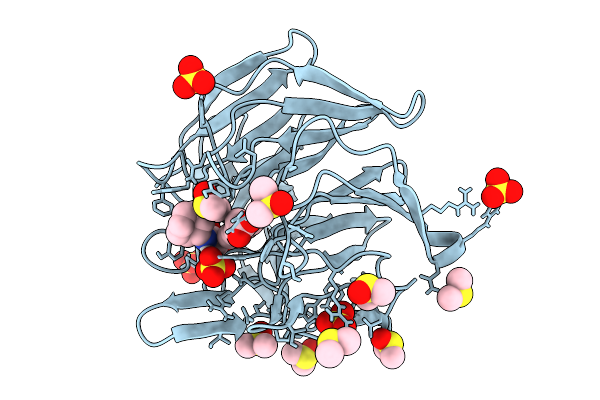 |
Crystal Structure Of The Keap1 Kelch Domain In Complex With The Small Molecule Ucab#909 At 1.61 Angstrom Resolution
Organism: Mus musculus
Method: X-RAY DIFFRACTION Release Date: 2025-09-03 Classification: PEPTIDE BINDING PROTEIN Ligands: A1IX3, SO4, DMS, CL |
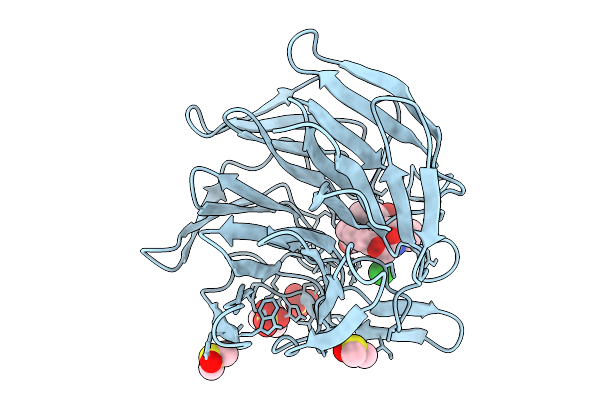 |
Crystal Structure Of The Keap1 Kelch Domain In Complex With The Small Molecule Ucab#985 At 1.65 Angstrom Resolution
Organism: Mus musculus
Method: X-RAY DIFFRACTION Release Date: 2025-09-03 Classification: PEPTIDE BINDING PROTEIN Ligands: A1IX4, SO4, DMS, CL |
 |
Crystal Structure Of The Keap1 Kelch Domain In Complex With The Small Molecule Ucab#1004 At 1.40 Angstrom Resolution
Organism: Mus musculus
Method: X-RAY DIFFRACTION Release Date: 2025-09-03 Classification: PEPTIDE BINDING PROTEIN Ligands: A1IXY, SO4, CL, DMS |
 |
Crystal Structure Of The Keap1 Kelch Domain In Complex With The Small Molecule Ucab#1010 At 1.50 Angstrom Resolution
Organism: Mus musculus
Method: X-RAY DIFFRACTION Release Date: 2025-09-03 Classification: PEPTIDE BINDING PROTEIN Ligands: A1IXZ, SO4, CL, DMS |
 |
Crystal Structure Of The Keap1 Kelch Domain In Complex With The Small Molecule Ucab#1032 At 1.61 Angstrom Resolution
Organism: Mus musculus
Method: X-RAY DIFFRACTION Release Date: 2025-09-03 Classification: PEPTIDE BINDING PROTEIN Ligands: A1IX0, SO4, DMS |
 |
Crystal Structure Of The Keap1 Kelch Domain In Complex With The Small Molecule Ucab#1090 At 1.74 Angstrom Resolution
Organism: Mus musculus
Method: X-RAY DIFFRACTION Release Date: 2025-09-03 Classification: PEPTIDE BINDING PROTEIN Ligands: A1IX1, SO4, DMS |
 |
Crystal Structure Of 21A08Ap1-Fab In Complex With Human Pd-1 At 1.85 Angstrom Resolution
Organism: Mus musculus, Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-08-13 Classification: IMMUNE SYSTEM Ligands: EDO, DMS, PO4, NAG |
 |
The Structure Of Candida Albicans Phosphoglucose Isomerase In Complex With Fragments
Organism: Candida albicans
Method: X-RAY DIFFRACTION Release Date: 2025-07-30 Classification: ISOMERASE Ligands: PA5, A1IHU |
 |
The Structure Of Candida Albicans Phosphoglucose Isomerase In Complex With Fragments
Organism: Candida albicans
Method: X-RAY DIFFRACTION Release Date: 2025-07-30 Classification: ISOMERASE Ligands: A1IIB, PA5, CL |
 |
Organism: Streptococcus pyogenes
Method: X-RAY DIFFRACTION Release Date: 2025-07-30 Classification: OXIDOREDUCTASE Ligands: MPD, NAD, A1IH7 |
 |
The Structure Of Aspergillus Fumigatus Udp-Glcnac Pyrophosphorylase In Complex With A Fragment
Organism: Aspergillus fumigatus
Method: X-RAY DIFFRACTION Release Date: 2025-07-30 Classification: TRANSFERASE Ligands: GN1, A1IIG, 71G, MG, CL |
 |
The Structure Of Aspergillus Fumigatus Udp-Glcnac Pyrophosphorylase In Complex With A Fragment
Organism: Aspergillus fumigatus
Method: X-RAY DIFFRACTION Release Date: 2025-07-30 Classification: TRANSFERASE Ligands: A1IHU, GN1 |
 |
The Structure Of Aspergillus Fumigatus Udp-Glcnac Pyrophosphorylase In Complex With A Fragment
Organism: Aspergillus fumigatus
Method: X-RAY DIFFRACTION Release Date: 2025-07-30 Classification: TRANSFERASE Ligands: GN1, YRL, MG |

