Search Count: 85
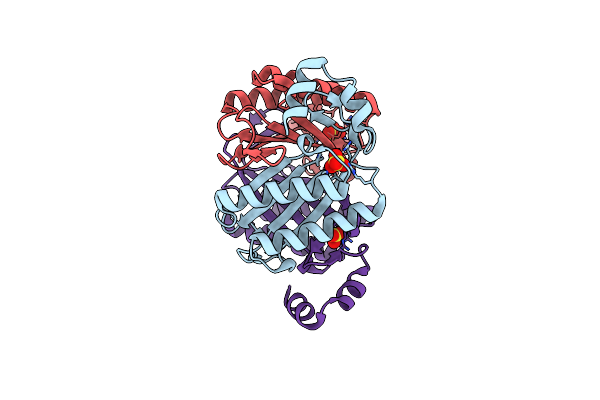 |
Crystal Structure Of N1, A Member Of Cis-3-Chloroacrylic Acid Dehalogenase (Cis-Caad) Family
Organism: Corynebacterium halotolerans yim 70093 = dsm 44683
Method: X-RAY DIFFRACTION Resolution:2.45 Å Release Date: 2021-06-02 Classification: HYDROLASE Ligands: SO4 |
 |
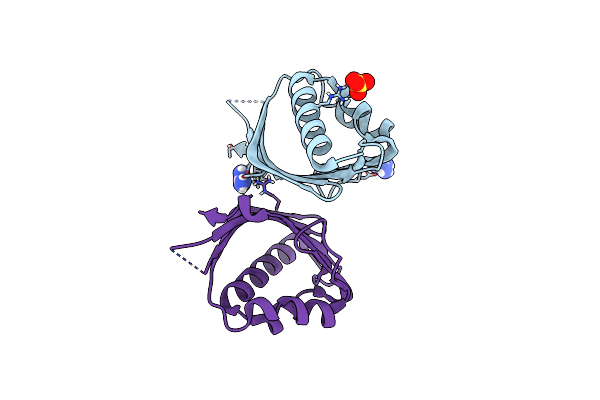
Crystal Structure Of N2, A Member Of 4-Oxalocrotonate Tautomerase (4-Ot) Family
Organism: Gammaproteobacteria bacterium sg8_31
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2021-06-02 Classification: ISOMERASE |
 |
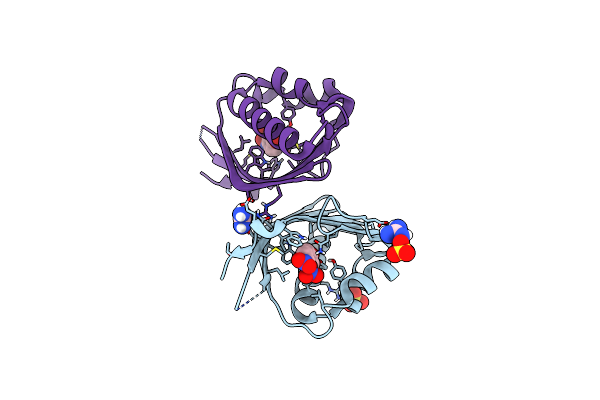
Crystal Structure Of Ketosteroid Isomerase From Mycobacterium Hassiacum (Mhksi)
Organism: Mycobacterium hassiacum (strain dsm 44199 / cip 105218 / jcm 12690 / 3849)
Method: X-RAY DIFFRACTION Resolution:1.57 Å Release Date: 2020-05-27 Classification: ISOMERASE Ligands: SO4, GAI |
 |
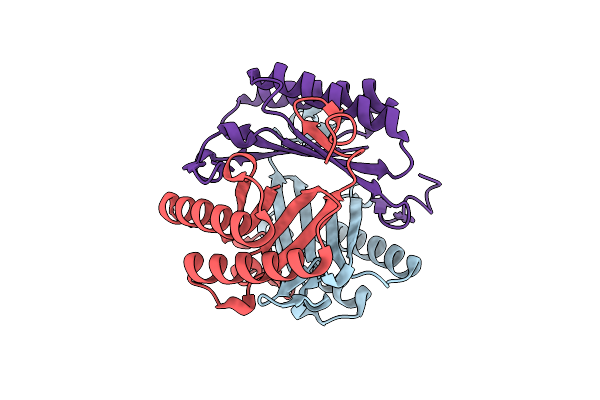
Crystal Structure Of Ketosteroid Isomerase D38N Mutant From Mycobacterium Hassiacum (Mhksi) Bound To 3,4-Dinitrophenol
Organism: Mycobacterium hassiacum (strain dsm 44199 / cip 105218 / jcm 12690 / 3849)
Method: X-RAY DIFFRACTION Resolution:1.25 Å Release Date: 2020-05-27 Classification: ISOMERASE Ligands: DNX, SO4, GAI |
 |
Organism: Burkholderia sp. rpe67
Method: X-RAY DIFFRACTION Resolution:1.83 Å Release Date: 2020-04-15 Classification: ISOMERASE |
 |
Organism: Caballeronia arationis
Method: X-RAY DIFFRACTION Resolution:1.39 Å Release Date: 2020-04-15 Classification: ISOMERASE Ligands: GOL |
 |
Organism: Bordetella trematum
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2020-04-15 Classification: ISOMERASE |
 |
Organism: Advenella mimigardefordensis dpn7
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2020-04-15 Classification: ISOMERASE |
 |
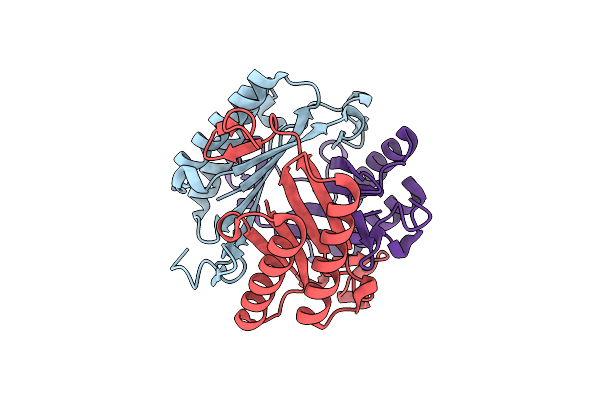
Organism: Burkholderia lata (strain atcc 17760 / dsm 23089 / lmg 22485 / ncimb 9086 / r18194 / 383)
Method: X-RAY DIFFRACTION Resolution:1.87 Å Release Date: 2020-02-26 Classification: HYDROLASE Ligands: GOL |
 |
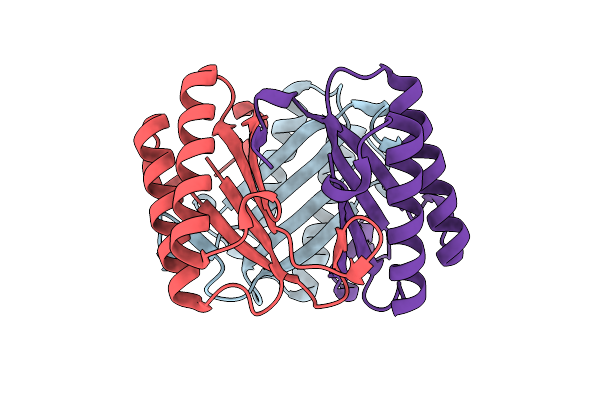
Organism: Burkholderia lata (strain atcc 17760 / dsm 23089 / lmg 22485 / ncimb 9086 / r18194 / 383)
Method: X-RAY DIFFRACTION Resolution:1.49 Å Release Date: 2017-12-06 Classification: HYDROLASE |
 |
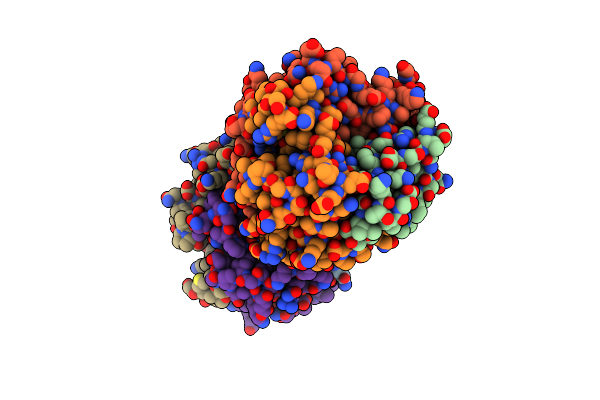
Organism: Pseudomonas sp. uw4
Method: X-RAY DIFFRACTION Resolution:2.57 Å Release Date: 2017-11-22 Classification: HYDROLASE |
 |
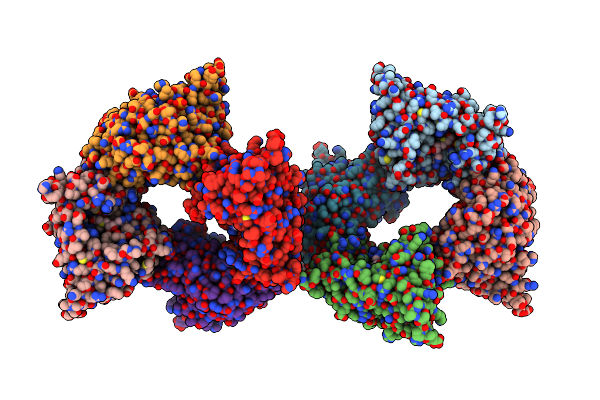
Organism: Pusillimonas sp. (strain t7-7)
Method: X-RAY DIFFRACTION Resolution:1.98 Å Release Date: 2017-11-22 Classification: HYDROLASE |
 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2014-09-17 Classification: TRANSFERASE Ligands: GEK, PO4 |
 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.64 Å Release Date: 2014-09-17 Classification: TRANSFERASE Ligands: PO4, GEK |
 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.62 Å Release Date: 2014-09-17 Classification: TRANSFERASE Ligands: SO4 |
 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2012-10-10 Classification: TRANSFERASE Ligands: SO4, GEK |
 |
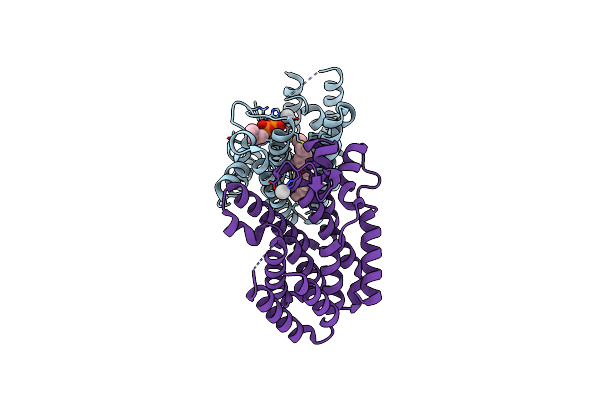
Crystal Structure Of An Enolase (Mandalate Racemase Subgroup, Target Efi-502101) From Pelagibaca Bermudensis Htcc2601, With Bound Mg And L-4-Hydroxyproline Betaine (Betonicine)
Organism: Pelagibaca bermudensis
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2012-10-10 Classification: ISOMERASE Ligands: MG, NI, 0XW, IOD, MPD |
 |
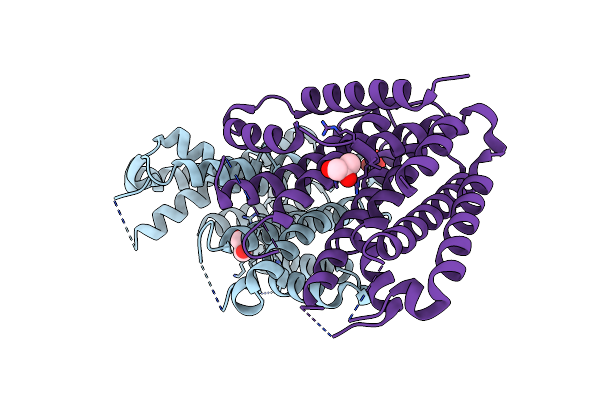
Crystal Structure Of Isoprenoid Synthase A3Mx09 (Target Efi-501993) From Pyrobaculum Calidifontis
Organism: Pyrobaculum calidifontis
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2012-07-11 Classification: TRANSFERASE Ligands: GER, UNX, UNL |
 |
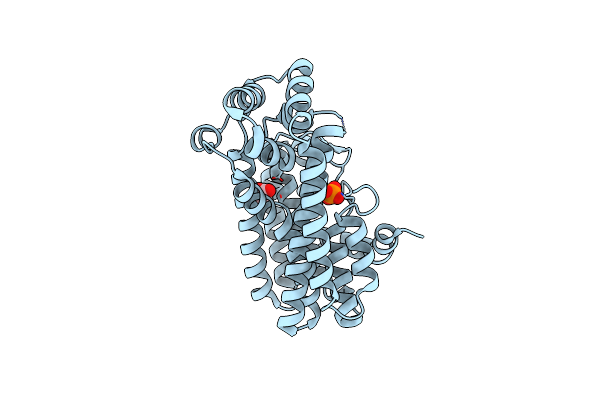
Crystal Structure Of A Putative Farnesyl-Diphosphate Synthase From Marinomonas Sp. Med121 (Target Efi-501980)
Organism: Marinomonas sp. med121
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2012-06-13 Classification: TRANSFERASE Ligands: SO4, GOL, CL |
 |
Crystal Structure Of Isoprenoid Synthase A3Msh1 (Target Efi-501992) From Pyrobaculum Calidifontis
Organism: Pyrobaculum calidifontis
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2012-02-08 Classification: TRANSFERASE Ligands: PO4, CL, ACT |

