Search Count: 270
 |
Organism: Enterobacteria phage t4
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2010-01-19 Classification: HYDROLASE Ligands: BME |
 |
Sequential Reorganization Of Beta-Sheet Topology By Insertion Of A Single Strand
Organism: Enterobacteria phage t4
Method: X-RAY DIFFRACTION Resolution:3.00 Å Release Date: 2009-10-20 Classification: HYDROLASE Ligands: SO4 |
 |
T4 Lysozyme M102E/L99A Mutant With Buried Charge In Apolar Cavity--Apo Structure
Organism: Enterobacteria phage t4
Method: X-RAY DIFFRACTION Resolution:1.45 Å Release Date: 2009-08-25 Classification: HYDROLASE Ligands: BME, CO3, TAM, SO4 |
 |
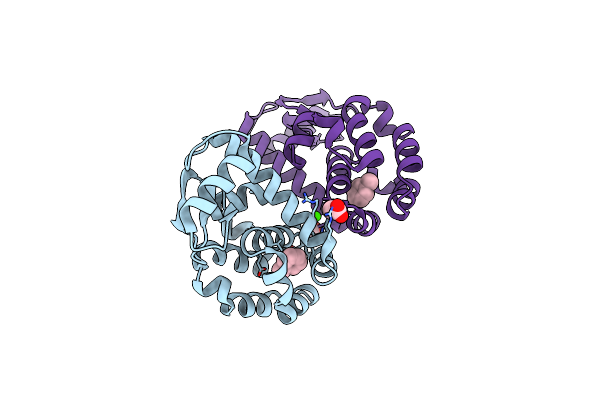
T4 Lysozyme M102E/L99A Mutant With Buried Charge In Apolar Cavity--Benzene Binding
Organism: Enterobacteria phage t4
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2009-08-25 Classification: HYDROLASE Ligands: BNZ, BME, CO3, EPE, SO4 |
 |
T4 Lysozyme M102E/L99A Mutant With Buried Charge In Apolar Cavity--Toluene Binding
Organism: Enterobacteria phage t4
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2009-08-25 Classification: HYDROLASE Ligands: MBN, CA, CL, ACT |
 |
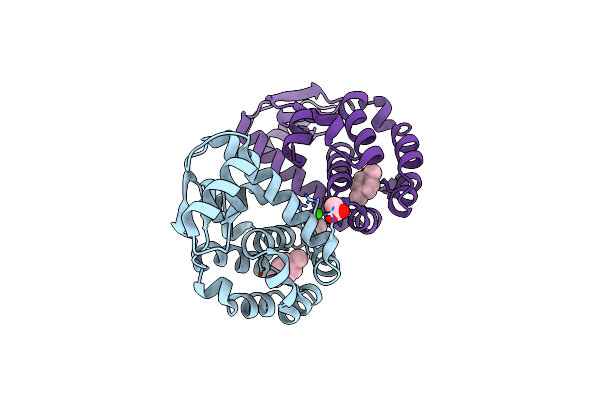
T4 Lysozyme M102E/L99A Mutant With Buried Charge In Apolar Cavity--Ethylbenzene Binding
Organism: Enterobacteria phage t4
Method: X-RAY DIFFRACTION Resolution:2.07 Å Release Date: 2009-08-25 Classification: HYDROLASE Ligands: PYJ, ACT, CA, CL |
 |
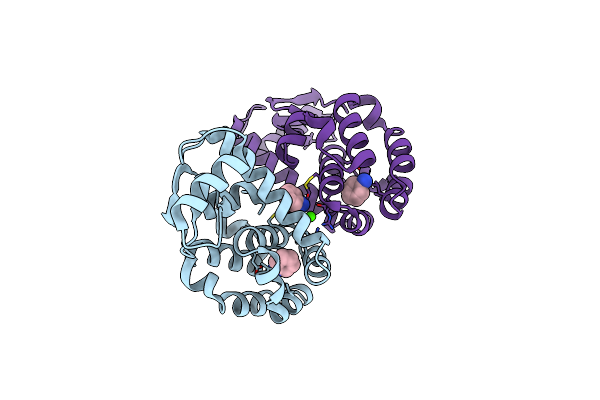
T4 Lysozyme M102E/L99A Mutant With Buried Charge In Apolar Cavity--P-Xylene Binding
Organism: Enterobacteria phage t4
Method: X-RAY DIFFRACTION Resolution:2.24 Å Release Date: 2009-08-25 Classification: HYDROLASE Ligands: PXY, CL, ACT, CA |
 |
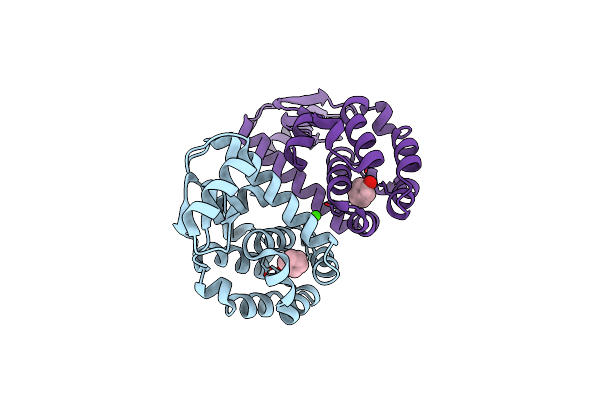
T4 Lysozyme M102E/L99A Mutant With Buried Charge In Apolar Cavity--Aniline Binding
Organism: Enterobacteria phage t4
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2009-08-25 Classification: HYDROLASE Ligands: ANL, CA, CL |
 |
T4 Lysozyme M102E/L99A Mutant With Buried Charge In Apolar Cavity--Phenol Binding
Organism: Enterobacteria phage t4
Method: X-RAY DIFFRACTION Resolution:2.16 Å Release Date: 2009-08-25 Classification: HYDROLASE Ligands: IPH, CL, CA |
 |
T4 Lysozyme M102E/L99A Mutant With Buried Charge In Apolar Cavity--Pyridine Binding
Organism: Enterobacteria phage t4
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2009-08-25 Classification: HYDROLASE Ligands: 0PY, CO3, MG, CL |
 |
Contributions Of All 20 Amino Acids At Site 96 To The Stability And Structure Of T4 Lysozyme
Organism: Bacteriophage t4
Method: X-RAY DIFFRACTION Resolution:1.77 Å Release Date: 2009-02-17 Classification: HYDROLASE Ligands: CL, BME |
 |
Organism: Bacteriophage t4
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2009-02-17 Classification: HYDROLASE Ligands: CL, BME |
 |
Organism: Bacteriophage t4
Method: X-RAY DIFFRACTION Resolution:1.67 Å Release Date: 2009-02-17 Classification: HYDROLASE Ligands: CL, BME |
 |
Organism: Bacteriophage t4
Method: X-RAY DIFFRACTION Resolution:1.99 Å Release Date: 2009-02-17 Classification: HYDROLASE Ligands: CL, BME |
 |
Organism: Bacteriophage t4
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2009-02-17 Classification: HYDROLASE Ligands: CL, BME |
 |
Organism: Bacteriophage t4
Method: X-RAY DIFFRACTION Resolution:1.68 Å Release Date: 2009-02-17 Classification: HYDROLASE Ligands: CL, BME |
 |
Bacteriophage T4 Lysozyme Mutant D89A In Wildtype Background At Room Temperature
Organism: Bacteriophage t4
Method: X-RAY DIFFRACTION Resolution:1.84 Å Release Date: 2009-02-17 Classification: HYDROLASE Ligands: CL, BME |
 |
Contribution Of All 20 Amino Acids At Site 96 To The Stability And Structure Of T4 Lysozyme
Organism: Bacteriophage t4
Method: X-RAY DIFFRACTION Resolution:1.64 Å Release Date: 2009-02-17 Classification: HYDROLASE Ligands: K, CL, BME |
 |
Contributions Of All 20 Amino Acids At Site 96 To Stability And Structure Of T4 Lysozyme
Organism: Bacteriophage t4
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2009-02-17 Classification: HYDROLASE Ligands: CL, BME |
 |
Contributions Of All 20 Amino Acids At Site 96 To The Stability And Structure Of T4 Lysozyme
Organism: Bacteriophage t4
Method: X-RAY DIFFRACTION Resolution:1.68 Å Release Date: 2009-02-17 Classification: HYDROLASE Ligands: CL, K, BME |

