Search Count: 17
 |
Crystal Structure Of Mutant Nonpro1 Tautomerase Superfamily Member Nj7-V1P In Complex With 3-Bromopropiolate Inhibitor
Organism: Nostoc sp.
Method: X-RAY DIFFRACTION Resolution:2.09 Å Release Date: 2025-03-05 Classification: ISOMERASE/INHIBITOR Ligands: A1AWY |
 |
Crystal Structure Of Mutant Nonpro1 Tautomerase Superfamily Member 8U6-S1P In Complex With 3-Bromopropiolate Inhibitor
Organism: Sulfurovum
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2025-03-05 Classification: ISOMERASE/INHIBITOR Ligands: A1AWY |
 |
Crystal Structure Of Cf, A Heterohexamer Of The 4-Oxalocrotonate Tautomerase (4-Ot) Family
Organism: Herbaspirillum
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2023-12-06 Classification: ISOMERASE |
 |
Crystal Structure Of Yr, A Heterohexamer Of The 4-Oxalocrotonate Tautomerase (4-Ot) Family
Organism: Herbaspirillum
Method: X-RAY DIFFRACTION Resolution:2.04 Å Release Date: 2023-12-06 Classification: ISOMERASE |
 |
Organism: Herbaspirillum
Method: X-RAY DIFFRACTION Resolution:2.29 Å Release Date: 2023-12-06 Classification: ISOMERASE |
 |
Structural, Kinetic, And Mechanistic Analysis Of The Wild-Type And Inactivated Malonate Semialdehyde Decarboxylase: A Structural Basis For The Decarboxylase And Hydratase Activities
Organism: Pseudomonas pavonaceae
Method: X-RAY DIFFRACTION Resolution:2.35 Å Release Date: 2022-12-21 Classification: ISOMERASE |
 |
Crystal Structure Of N1, A Member Of Cis-3-Chloroacrylic Acid Dehalogenase (Cis-Caad) Family
Organism: Corynebacterium halotolerans yim 70093 = dsm 44683
Method: X-RAY DIFFRACTION Resolution:2.45 Å Release Date: 2021-06-02 Classification: HYDROLASE Ligands: SO4 |
 |
Crystal Structure Of N2, A Member Of 4-Oxalocrotonate Tautomerase (4-Ot) Family
Organism: Gammaproteobacteria bacterium sg8_31
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2021-06-02 Classification: ISOMERASE |
 |
Organism: Burkholderia lata (strain atcc 17760 / dsm 23089 / lmg 22485 / ncimb 9086 / r18194 / 383)
Method: X-RAY DIFFRACTION Resolution:1.87 Å Release Date: 2020-02-26 Classification: HYDROLASE Ligands: GOL |
 |
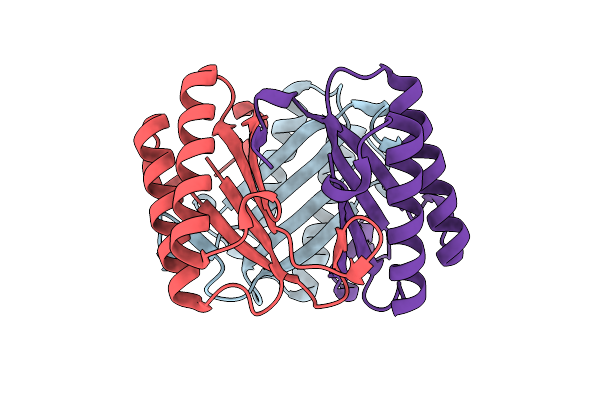
Organism: Burkholderia lata (strain atcc 17760 / dsm 23089 / lmg 22485 / ncimb 9086 / r18194 / 383)
Method: X-RAY DIFFRACTION Resolution:1.49 Å Release Date: 2017-12-06 Classification: HYDROLASE |
 |
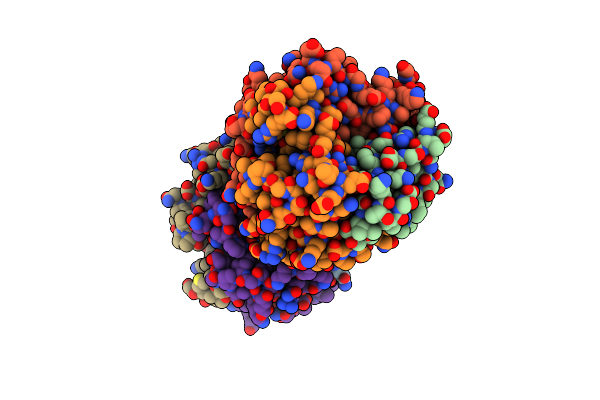
Organism: Pseudomonas sp. uw4
Method: X-RAY DIFFRACTION Resolution:2.57 Å Release Date: 2017-11-22 Classification: HYDROLASE |
 |
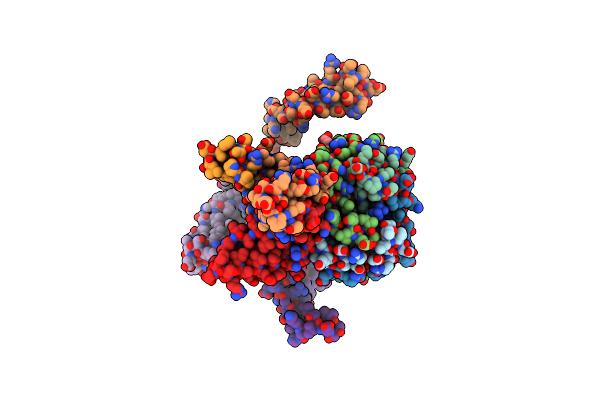
Organism: Pusillimonas sp. (strain t7-7)
Method: X-RAY DIFFRACTION Resolution:1.98 Å Release Date: 2017-11-22 Classification: HYDROLASE |
 |
Organism: Pseudomonas putida
Method: X-RAY DIFFRACTION Resolution:2.71 Å Release Date: 2016-03-09 Classification: ISOMERASE |
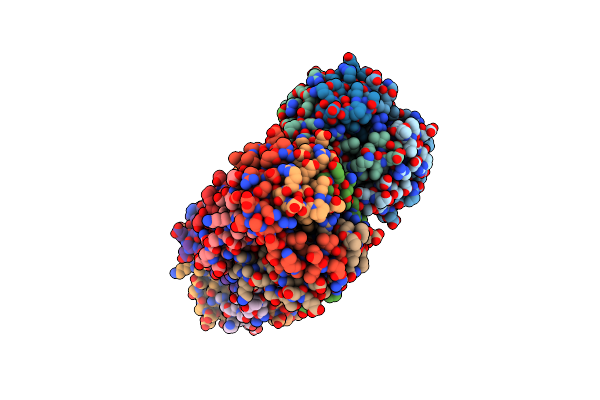 |
Crystal Structure Of A 4-Oxalocrotonate Tautomerase Mutant In Complex With Nitrostyrene At 2.3 Angstrom
Organism: Pseudomonas putida
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2016-03-09 Classification: ISOMERASE Ligands: NS8 |
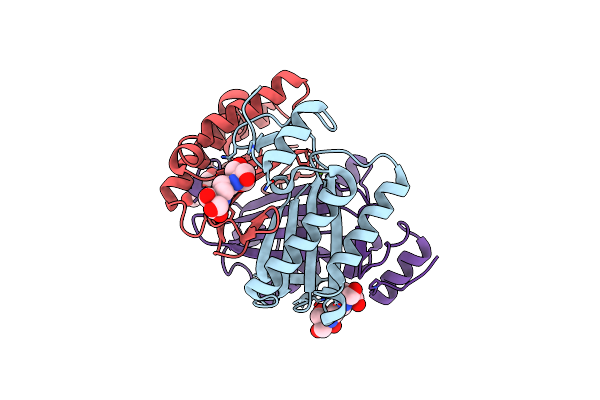 |
Crystal Structure Of Native Rhcc (Yp_702633.1) From Rhodococcus Jostii Rha1 At 1.78 Angstrom
Organism: Rhodococcus jostii rha1
Method: X-RAY DIFFRACTION Resolution:1.78 Å Release Date: 2015-02-25 Classification: ISOMERASE Ligands: B3P, MG, GOL |
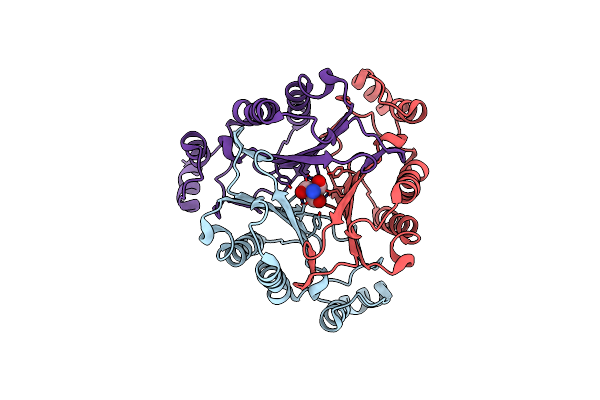 |
Crystal Structure Of D106A Mutant Of Rhcc (Yp_702633.1) From Rhodococcus Jostii Rha1 At 1.55 Angstrom
Organism: Rhodococcus jostii
Method: X-RAY DIFFRACTION Resolution:1.55 Å Release Date: 2015-02-25 Classification: ISOMERASE Ligands: TRS |
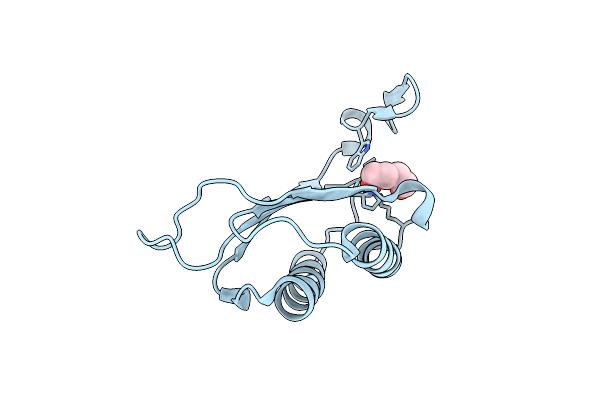 |
Crystal Structure Of Macrophage Migration Inhibitory Factor Homologue From Prochlorococcus Marinus
Organism: Prochlorococcus marinus
Method: X-RAY DIFFRACTION Resolution:1.64 Å Release Date: 2010-09-01 Classification: IMMUNE SYSTEM Ligands: PEG |

