Search Count: 20
 |
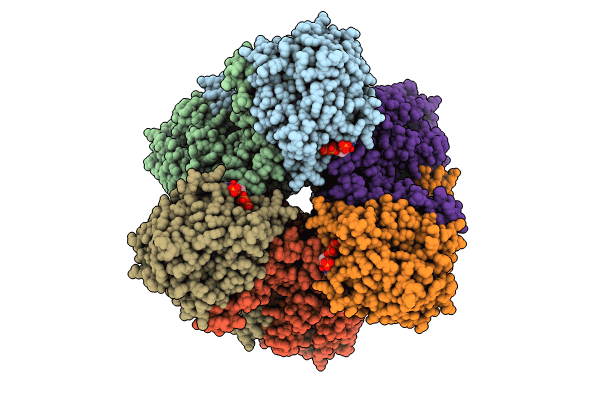
Cryo-Em Structure Of Active Phthaloyl-Coa Decarboxylase (Pcd) Complex With Prfmn Bound
Organism: Thauera chlorobenzoica
Method: ELECTRON MICROSCOPY Release Date: 2024-07-10 Classification: FLAVOPROTEIN Ligands: BYN, FE, K |
 |
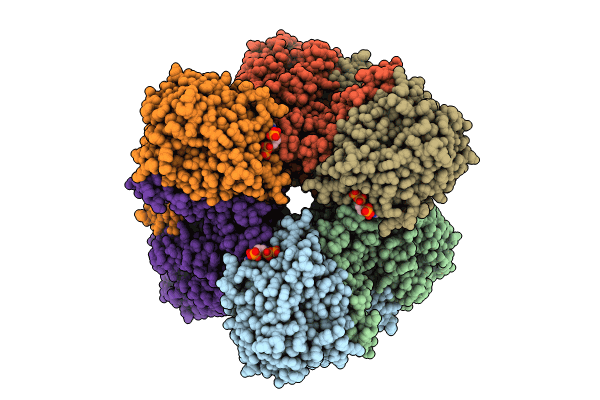
Cryo-Em Structure Of Phthaloyl-Coa Decarboxylase (Pcd) Bound With Substrate Analog/Inhibitor, 2-Cn-Benzoyl-Coa
Organism: Thauera chlorobenzoica
Method: ELECTRON MICROSCOPY Release Date: 2024-06-05 Classification: FLAVOPROTEIN Ligands: BYN, WC8, FE, K, CA |
 |
Cryo-Em Structure Of Phthaloyl-Coa Decarboxylase (Pcd) Bound With Product, Benzoyl-Coa
Organism: Thauera chlorobenzoica
Method: ELECTRON MICROSCOPY Release Date: 2024-06-05 Classification: FLAVOPROTEIN Ligands: BYC, BYN, FE, CA, K |
 |
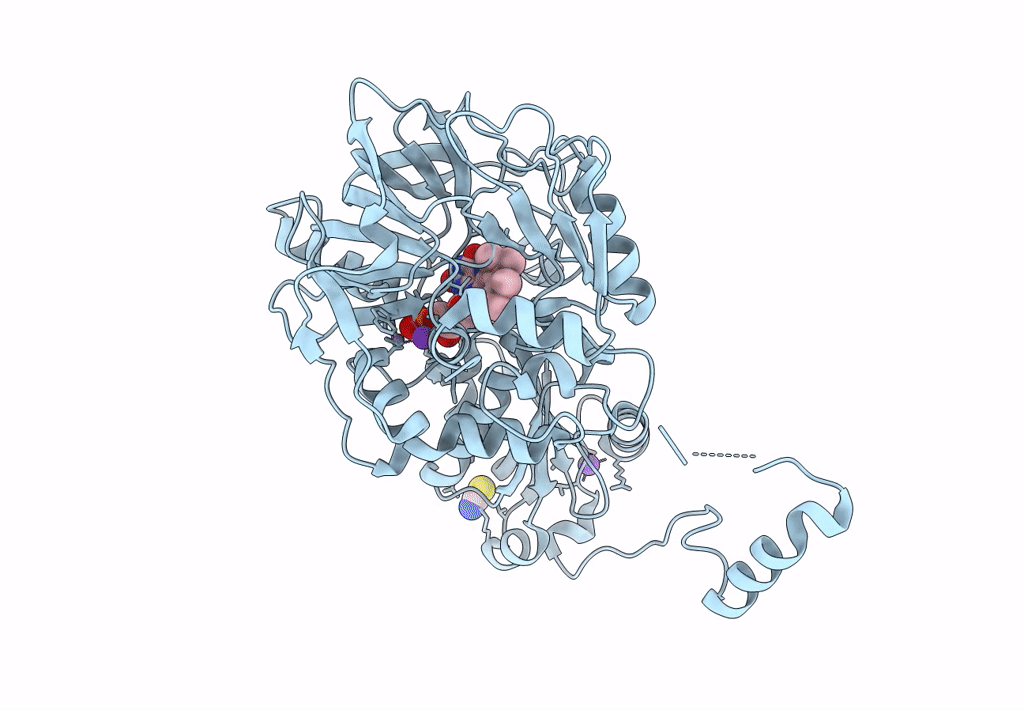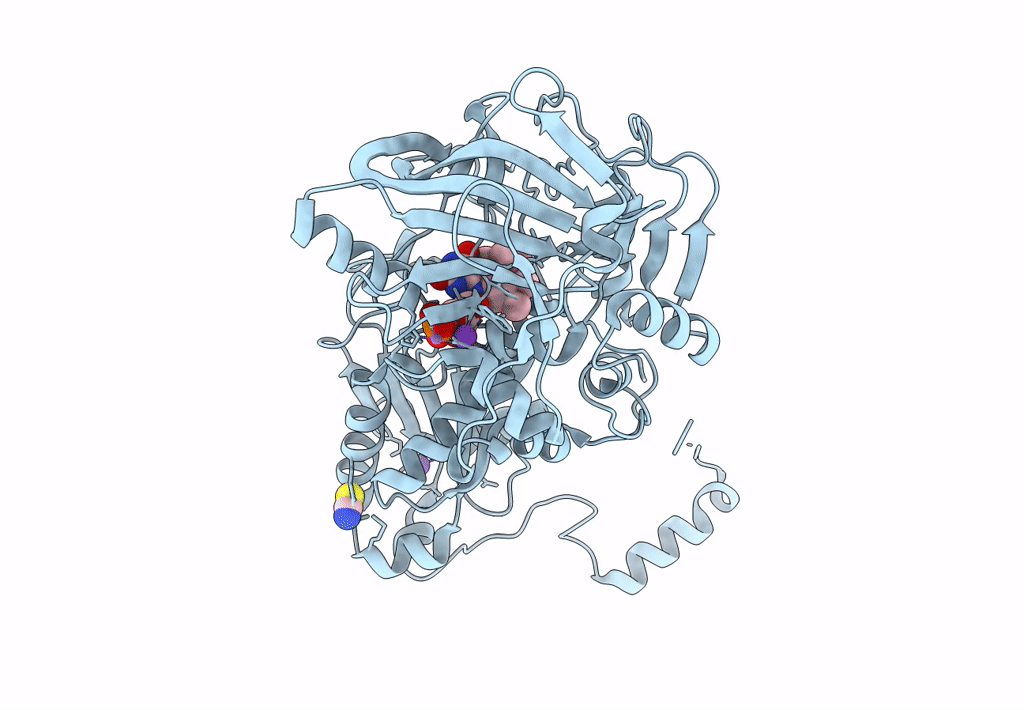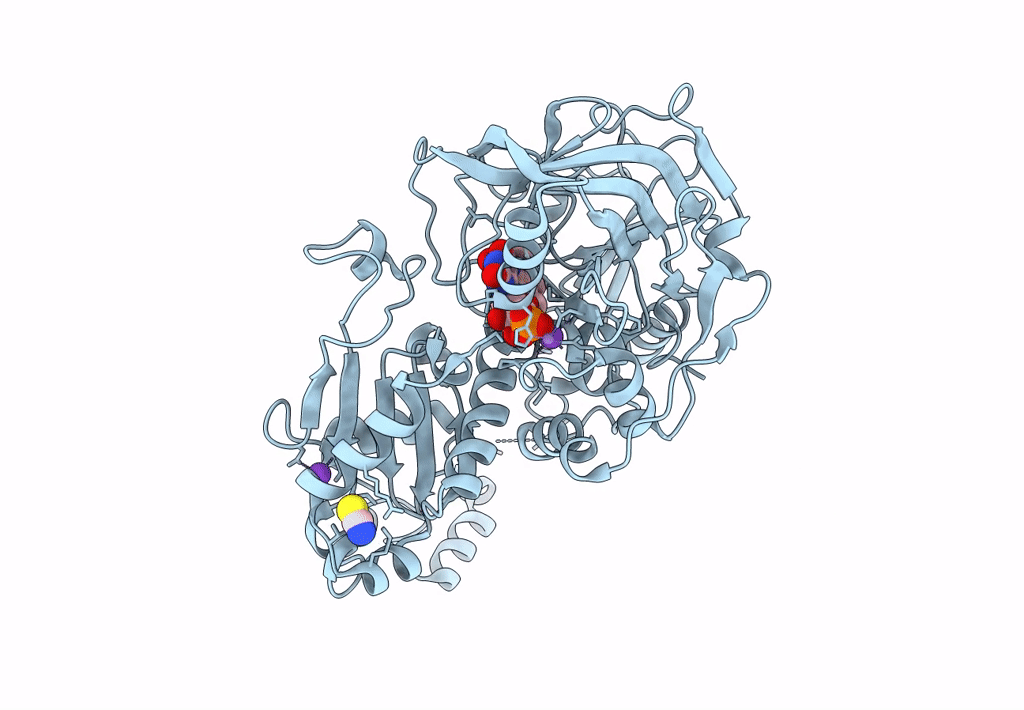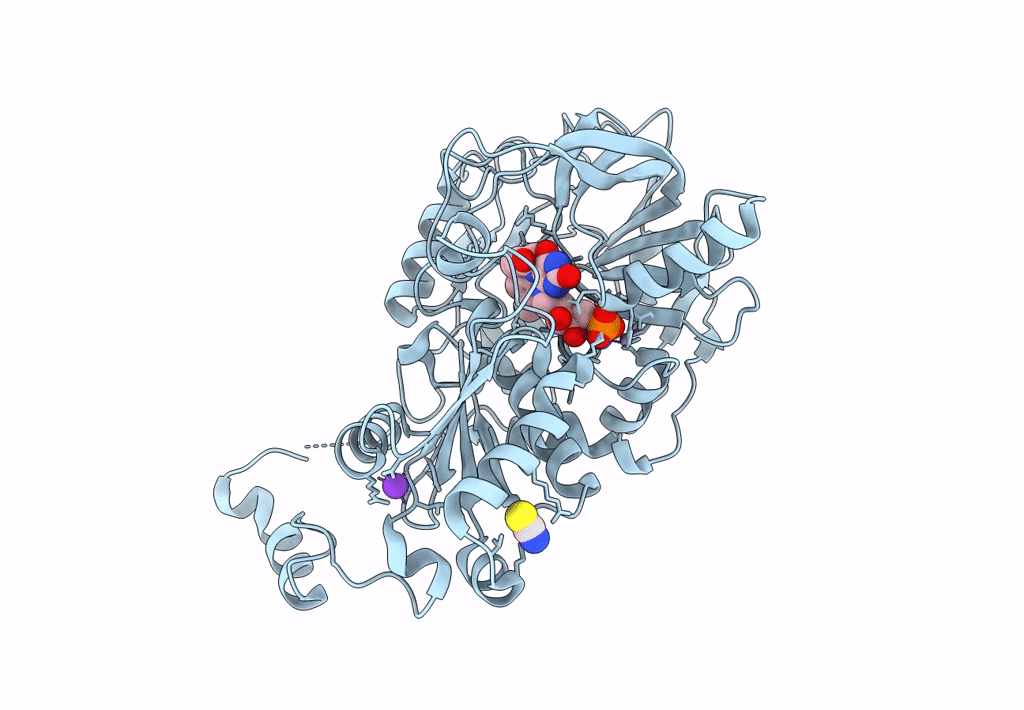
Aspergillus Niger Ferulic Acid Decarboxylase (Fdc) T40C-S315C (Db1) Variant In Complex With Prenylated Flavin Hydroxylated At The C1 Prime Position
Organism: Aspergillus niger cbs 513.88
Method: X-RAY DIFFRACTION Resolution:1.35 Å Release Date: 2023-09-06 Classification: LYASE Ligands: BYN, MN, K, SCN |
 |
Aspergillus Niger Ferulic Acid Decarboxylase (Fdc) S145C-P289C (Db2) Variant In Complex With Prenylated Flavin Hydroxylated At The C1 Prime Position
Organism: Aspergillus niger cbs 513.88
Method: X-RAY DIFFRACTION Resolution:1.37 Å Release Date: 2023-09-06 Classification: LYASE Ligands: BYN, MN, K, SCN |
 |
Organism: Hypocrea atroviridis (strain atcc 20476 / imi 206040)
Method: X-RAY DIFFRACTION Resolution:1.35 Å Release Date: 2021-08-04 Classification: LYASE Ligands: NO3, BYN, K, MN, ACT |
 |
Structure Of A. Niger Fdc T395M R435P P438W Variant (Anfdcii) In Complex With Prfmn
Organism: Aspergillus niger (strain cbs 513.88 / fgsc a1513)
Method: X-RAY DIFFRACTION Resolution:1.69 Å Release Date: 2021-08-04 Classification: LYASE Ligands: MN, K, BYN |
 |
Organism: Aspergillus niger
Method: X-RAY DIFFRACTION Resolution:1.07 Å Release Date: 2020-06-24 Classification: LIGASE Ligands: MN, K, BYN, FIV, PEG |
 |
Structure Of A. Niger Fdc I327S Variant In Complex With Benzothiophene 2 Carboxylic Acid
Organism: Aspergillus niger
Method: X-RAY DIFFRACTION Resolution:1.28 Å Release Date: 2020-06-24 Classification: LIGASE Ligands: MN, K, BYN, ND8 |
 |
Structure Of A. Niger Fdc I327S Variant In Complex With Indol-2-Carboxylic Acid
Organism: Aspergillus niger
Method: X-RAY DIFFRACTION Resolution:1.06 Å Release Date: 2020-06-24 Classification: LIGASE Ligands: MN, K, BYN, ICB |
 |
Organism: Aspergillus niger
Method: X-RAY DIFFRACTION Resolution:1.02 Å Release Date: 2020-06-24 Classification: LIGASE Ligands: MN, K, BYN, ND8 |
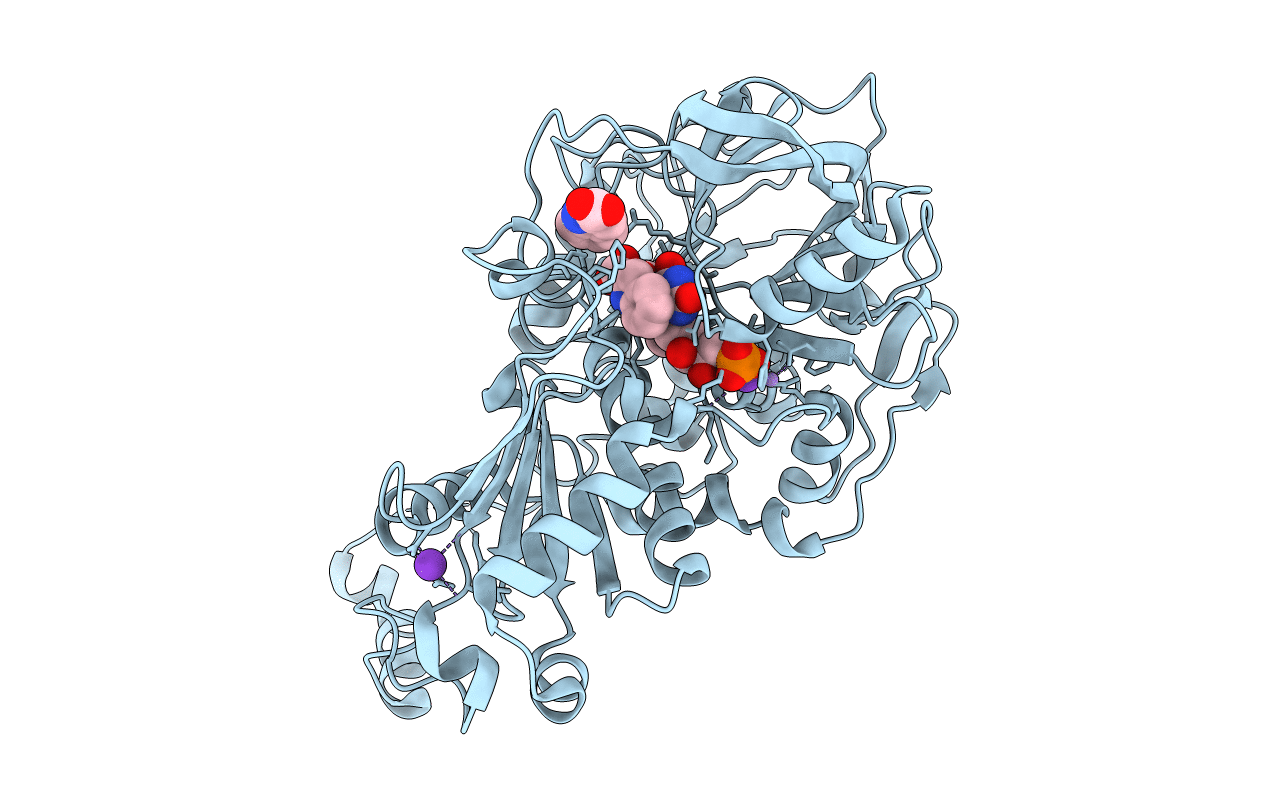 |
Organism: Aspergillus niger
Method: X-RAY DIFFRACTION Resolution:1.12 Å Release Date: 2020-06-24 Classification: LIGASE Ligands: MN, K, BYN, ICB |
 |
Aspergillus Niger Ferulic Acid Decarboxylase (Fdc) In Complex With Prfmn (Purified In Dark) And Alphafluorocinnamic Acid
Organism: Aspergillus niger
Method: X-RAY DIFFRACTION Resolution:1.13 Å Release Date: 2019-08-28 Classification: LYASE Ligands: MN, K, BYN, 4LW |
 |
Aspergillus Niger Ferulic Acid Decarboxylase (Fdc) In Complex With The Covalent Adduct Formed Between Prfmn Cofactor And Cinnamic Acid Following Decarboxylation (Int3)
Organism: Aspergillus niger
Method: X-RAY DIFFRACTION Resolution:1.24 Å Release Date: 2019-08-28 Classification: LYASE Ligands: MN, K, JSH, BYN, SYN |
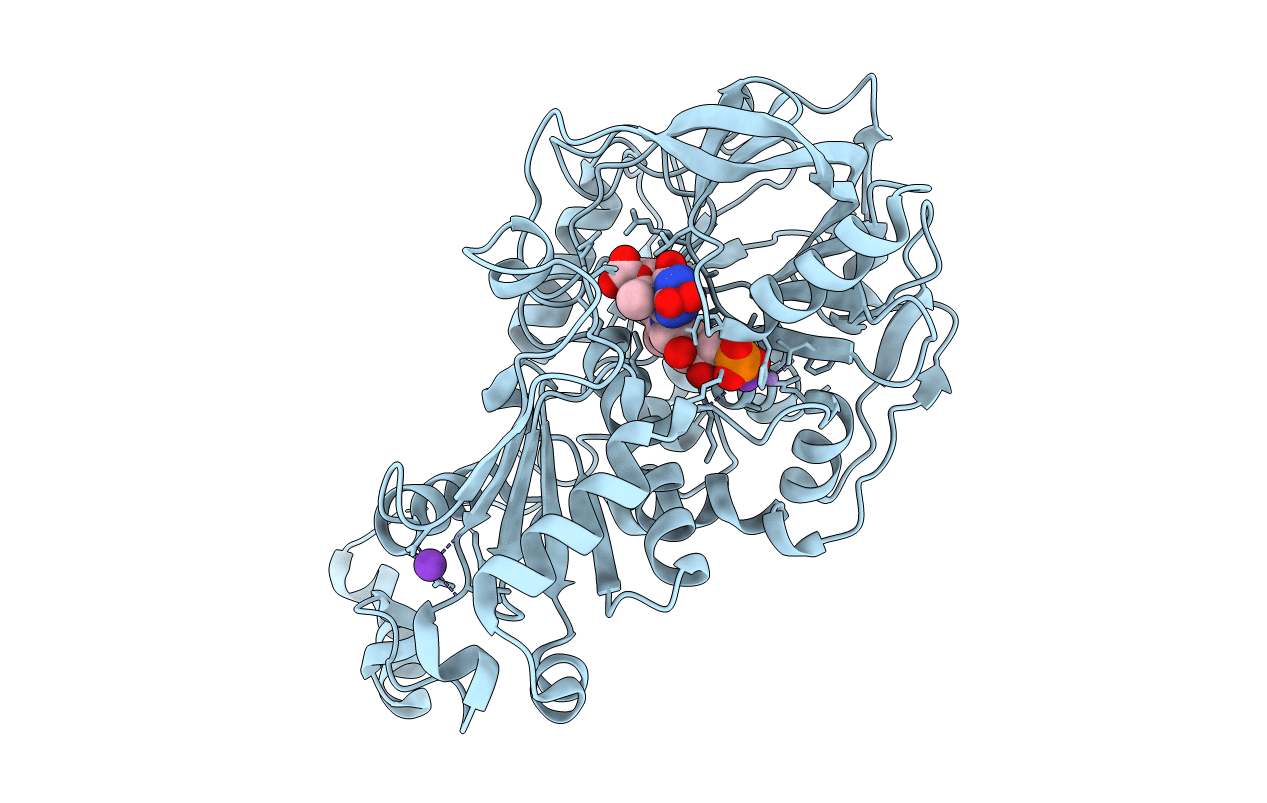 |
Aspergillus Niger Ferulic Acid Decarboxylase (Fdc) In Complex With The Covalent Adduct Formed Between Prfmn Cofactor And Butynoic Acid (Int1')
Organism: Aspergillus niger
Method: X-RAY DIFFRACTION Resolution:1.02 Å Release Date: 2019-08-28 Classification: LYASE Ligands: JRK, BYN, MN, K |
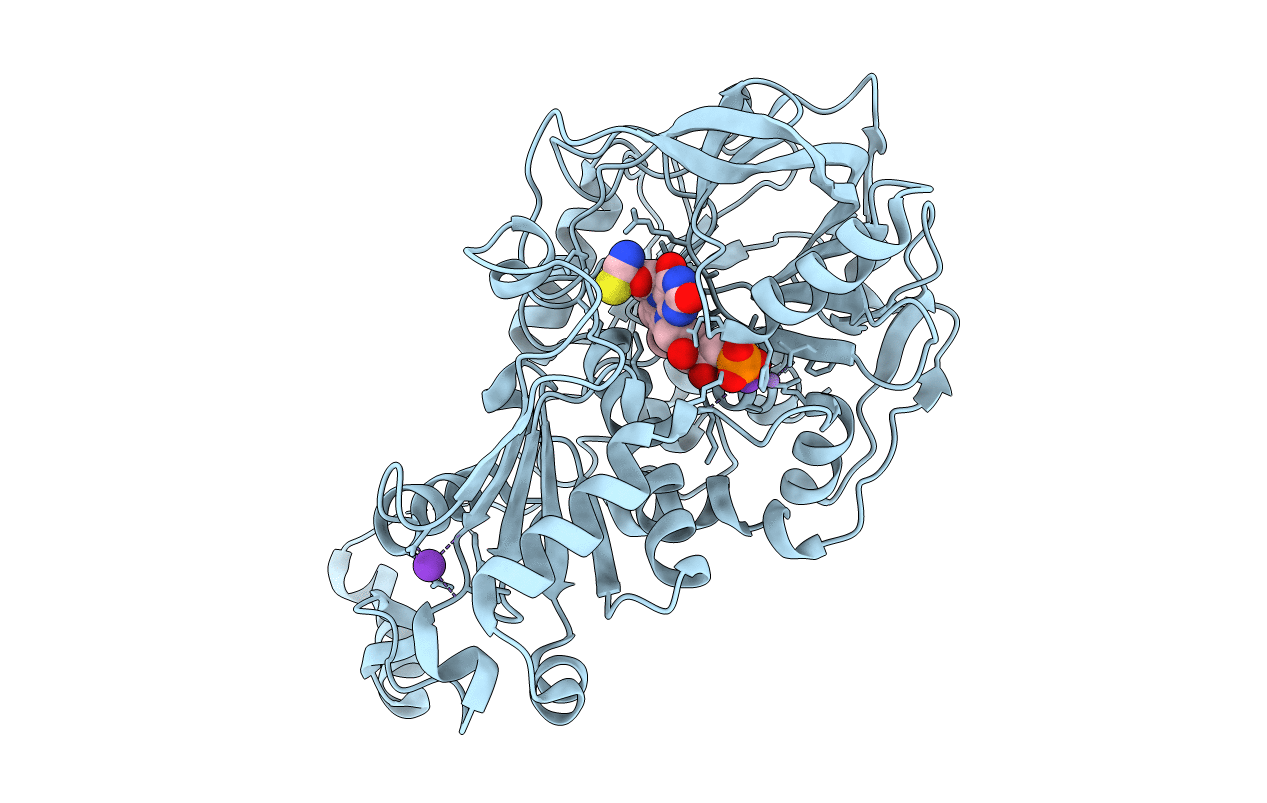 |
Organism: Aspergillus niger (strain cbs 513.88 / fgsc a1513)
Method: X-RAY DIFFRACTION Resolution:1.28 Å Release Date: 2017-12-20 Classification: LYASE Ligands: MN, K, BYN, SCN |
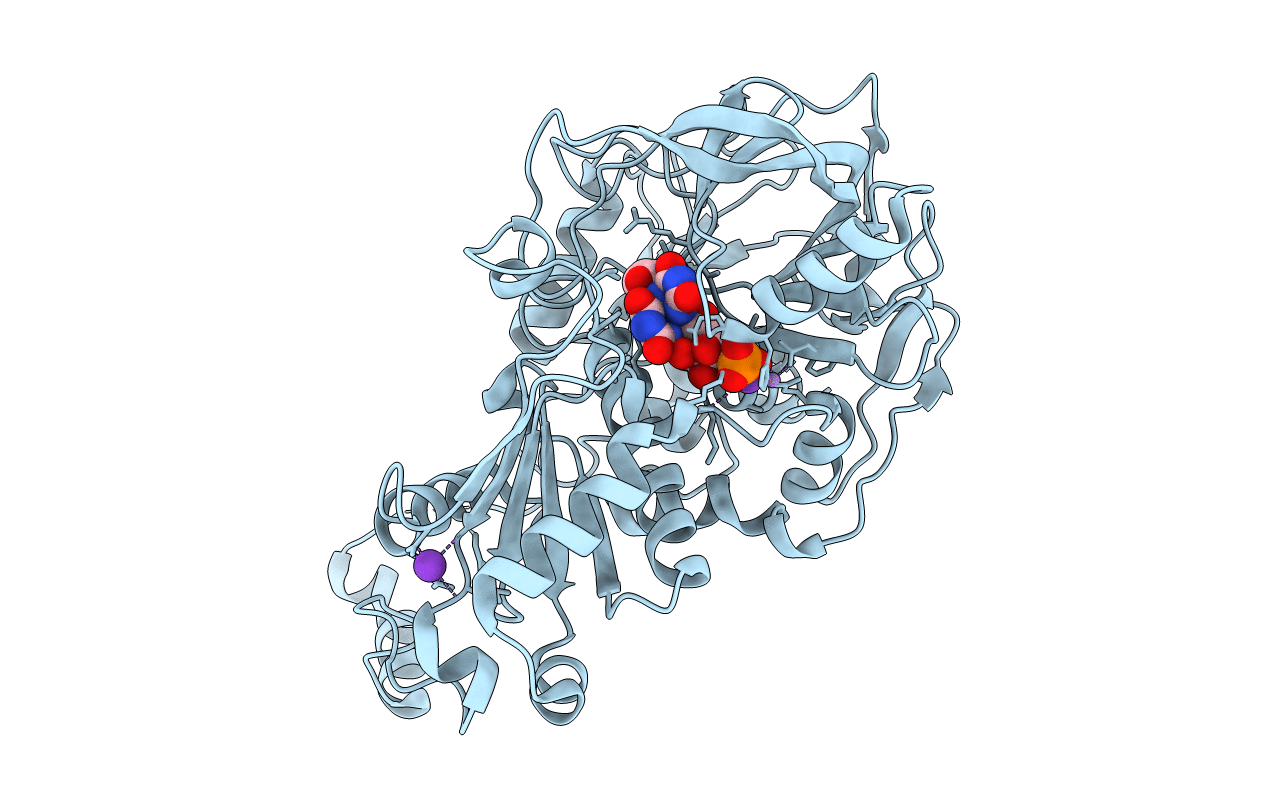 |
Structure Of E282Q A. Niger Fdc1 With Prfmn In The Hydroxylated And Ketimine Forms
Organism: Aspergillus niger (strain cbs 513.88 / fgsc a1513)
Method: X-RAY DIFFRACTION Resolution:1.10 Å Release Date: 2017-12-20 Classification: LYASE Ligands: MN, K, FZZ, BYN |
 |
Organism: Aspergillus niger (strain cbs 513.88 / fgsc a1513)
Method: X-RAY DIFFRACTION Resolution:1.03 Å Release Date: 2017-12-20 Classification: LYASE Ligands: MN, K, BYN, SCN |
 |
Organism: Aspergillus niger
Method: X-RAY DIFFRACTION Resolution:1.64 Å Release Date: 2017-12-20 Classification: LYASE Ligands: MN, K, BYN |
 |
Organism: Aspergillus niger (strain cbs 513.88 / fgsc a1513)
Method: X-RAY DIFFRACTION Resolution:1.19 Å Release Date: 2017-12-20 Classification: LYASE Ligands: MN, K, BYN |

