Planned Maintenance: Some services may turn out to be unavailable from 15th January, 2026 to 16th January, 2026. We apologize for the inconvenience!
Planned Maintenance: Some services may turn out to be unavailable from 15th January, 2026 to 16th January, 2026. We apologize for the inconvenience!
 |
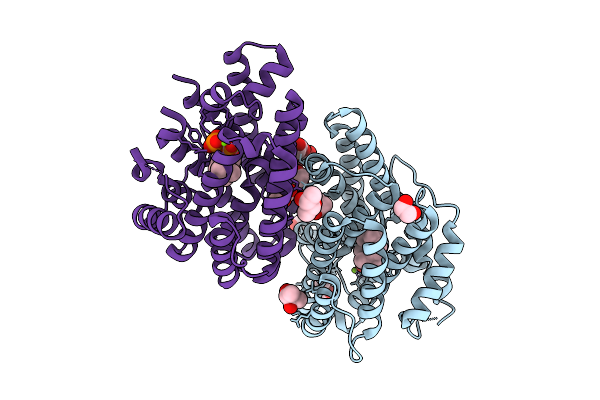
1-Epi-Cubenol Synthase From Nonomuraea Coxensis (Ncecs) In Complex With 2,3-Dhfpp
Organism: Nonomuraea coxensis
Method: X-RAY DIFFRACTION Release Date: 2025-12-24 Classification: LYASE Ligands: 3E9, MG, BU3, NA |
 |
T-Muurolol Synthase From Roseiflexus Castenholzii (Tms) In Complex With 2,3-Dhfpp [P4(3)2(1)2(1)]
Organism: Roseiflexus castenholzii
Method: X-RAY DIFFRACTION Release Date: 2025-12-24 Classification: LYASE Ligands: MG, 3E9, BU3, EDO, GOL |
 |
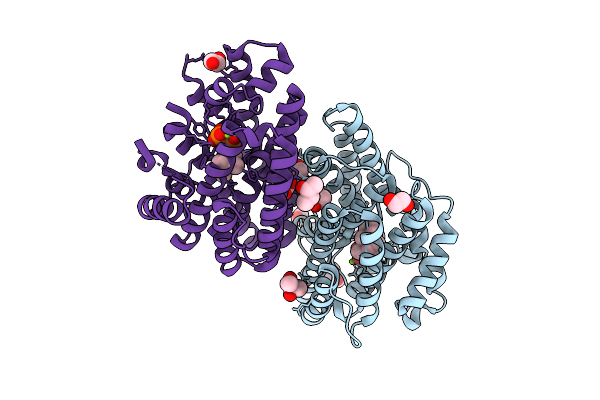
T-Muurolol Synthase From Roseiflexus Castenholzii (Tms) In Complex With Converted 1,2-Dhnpp (Compound 1)
Organism: Roseiflexus castenholzii
Method: X-RAY DIFFRACTION Release Date: 2025-12-24 Classification: LYASE Ligands: MG, POP, FAR, BU3, EDO |
 |
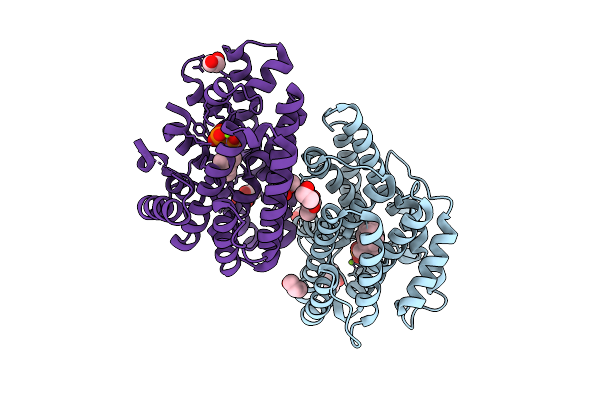
T-Muurolol Synthase From Roseiflexus Castenholzii (Tms) In Complex With Converted 10,11-Dhfpp (Compound 2+)
Organism: Roseiflexus castenholzii
Method: X-RAY DIFFRACTION Release Date: 2025-12-24 Classification: LYASE Ligands: MG, POP, A1IW0, BU3, EDO |
 |
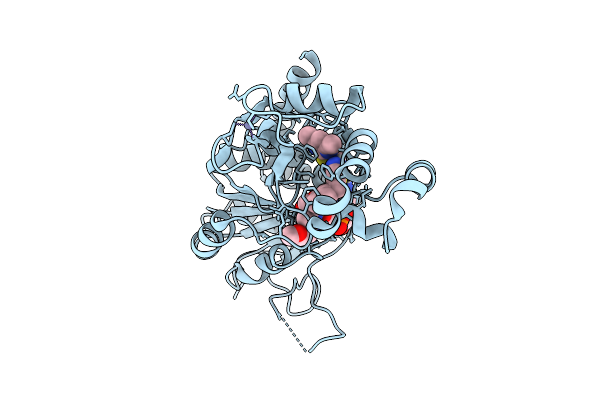
T-Muurolol Synthase From Roseiflexus Castenholzii (Tms) In Complex With Converted 6,7-Dhfpp (Compound 3+)
Organism: Roseiflexus castenholzii
Method: X-RAY DIFFRACTION Release Date: 2025-12-24 Classification: LYASE Ligands: MG, POP, A1IWZ, BU3, EDO, MES |
 |
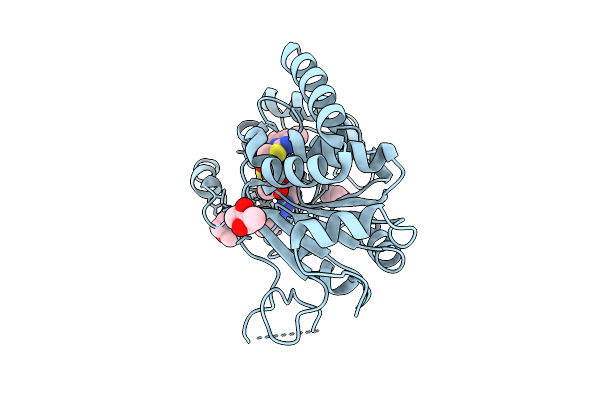
Crystal Structure Of Human Sirt2 In Complex With The Covalent Adduct Of Sirreal-Triazole Inhibitor Mz242 And Adp-Ribose
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-11-12 Classification: HYDROLASE Ligands: A1JK7, BU3, ZN |
 |
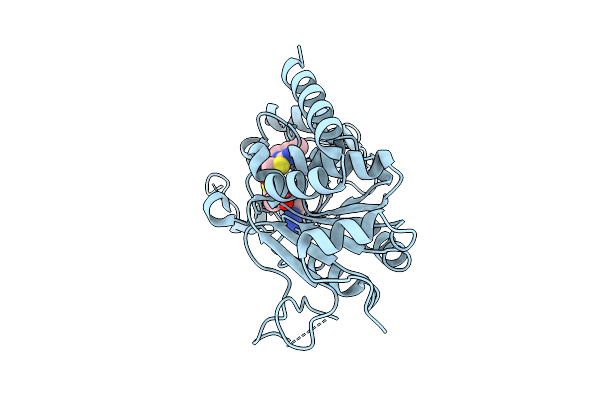
Crystal Structure Of Human Sirt2 In Complex With Sirreal-Triazole Inhibitor Sh10
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-11-12 Classification: HYDROLASE Ligands: ZN, BU3, EDO, A1JK0 |
 |
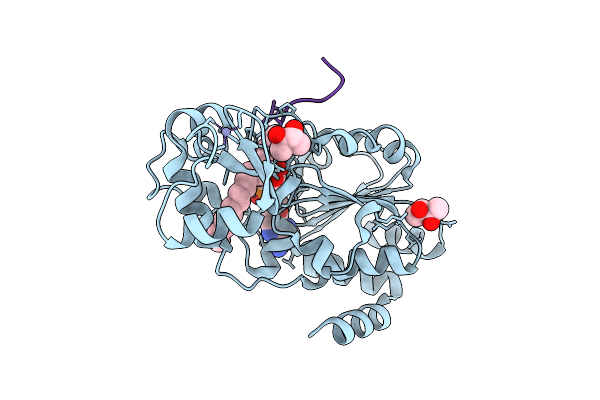
Crystal Structure Of Human Sirt2 In Complex With Sirreal-Triazole Inhibitor Lg023
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-11-12 Classification: HYDROLASE Ligands: ZN, A1JK1, BU3 |
 |
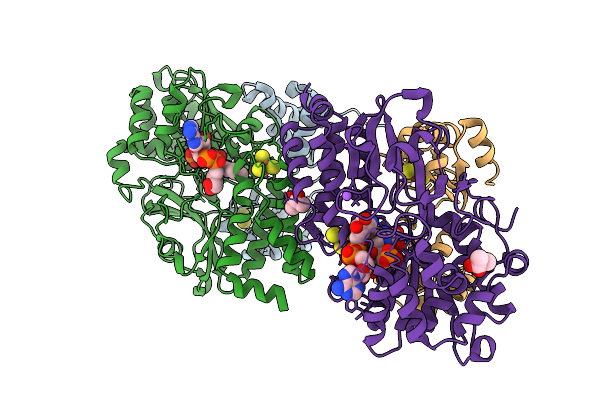
Crystal Structure Of Human Sirt3 In Complex With The Covalent Adduct Of Peptide Triazole Inhibitor Ltdi1 And Adp-Ribose
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-11-12 Classification: HYDROLASE Ligands: ZN, BU3, A1JK8 |
 |
Crystal Structure Of The Oxidized Respiratory Complex I Subunit Nuoef From Aquifex Aeolicus, Double Mutation V90P And V136M(Nuoe), Bound To Nad+
Organism: Aquifex aeolicus vf5
Method: X-RAY DIFFRACTION Release Date: 2025-11-05 Classification: OXIDOREDUCTASE Ligands: FES, SF4, FMN, NAD, NA, BU3, CL |
 |
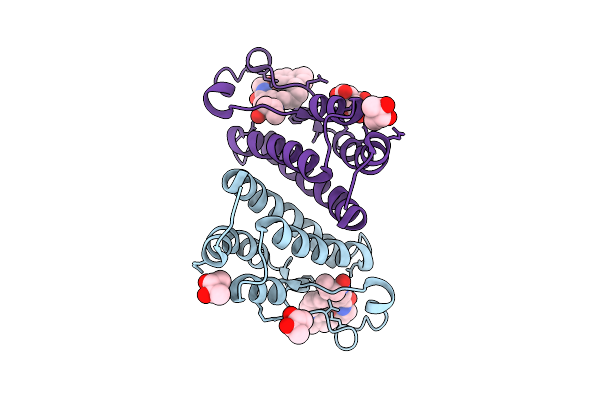
Crystal Structure Of The Leishmania Bromodomain Lmxbdf5.1 In Complex With Oxfbd06
Organism: Leishmania mexicana
Method: X-RAY DIFFRACTION Release Date: 2025-09-03 Classification: TRANSCRIPTION Ligands: A1IMI, BU3, K |
 |
Crystal Structure Of Crebbp Bromodomain In Complex With (R,E)-2-Methyl-1,2,3,5,10,11,17,18,21,22-Decahydro-4H,16H,20H-13,15-Etheno-7,25-(Metheno)[1,4]Diazepino[2,3-H]Pyrido[1,2-P][1]Oxa[4,16]Diazacyclononadecine-4,8(9H)-Dione
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-08-20 Classification: TRANSFERASE Ligands: BU3, A1IKL, CIT |
 |
Crystal Structure Of Crebbp Bromodomain In Complex With (R,E)-6-(5-(6-Methoxy-2,3-Dihydro-4H-Benzo[B][1,4]Oxazin-4-Yl)Pent-1-En-1-Yl)-4-Methyl-1,3,4,5-Tetrahydro-2H-Benzo[B][1,4]Diazepin-2-One
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-08-20 Classification: TRANSFERASE Ligands: A1IKI, BU3 |
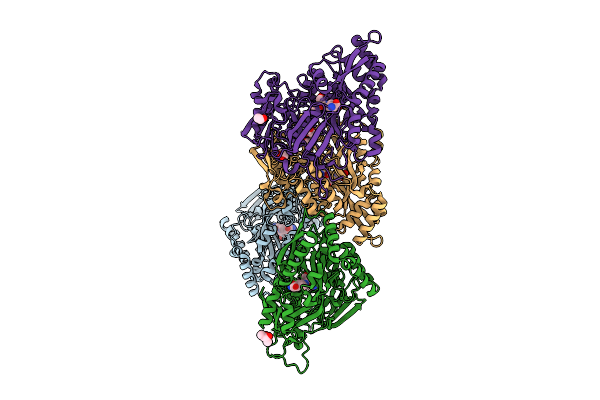 |
Organism: Escherichia coli, Synthetic construct
Method: X-RAY DIFFRACTION Release Date: 2025-08-13 Classification: TRANSPORT PROTEIN Ligands: A1JHR, BU3, EDO, GOL |
 |
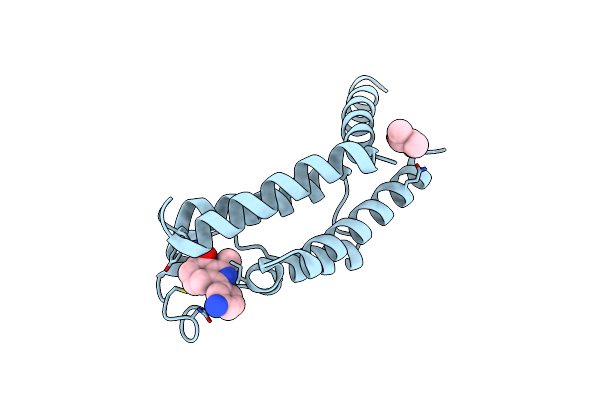
Organism: Legionella pneumophila str. corby
Method: X-RAY DIFFRACTION Release Date: 2025-06-18 Classification: HYDROLASE Ligands: A1IRL, BU3, GOL, ZN, SO4 |
 |
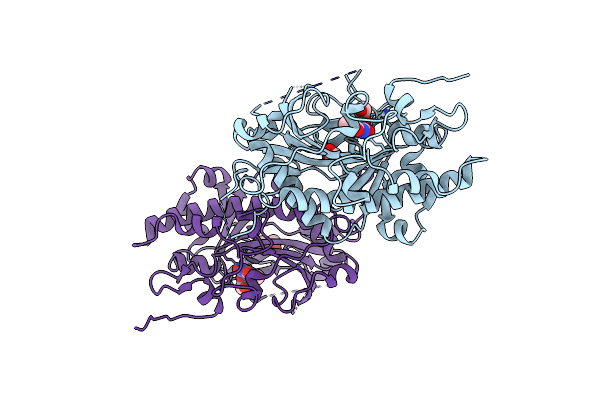
Crystal Structure Of The Plasmodium Falciparum Bromodomain Pfbdp1 In Complex With Mpm2
Organism: Plasmodium falciparum 3d7
Method: X-RAY DIFFRACTION Release Date: 2025-04-02 Classification: TRANSCRIPTION Ligands: BU3, I5K |
 |
Crystal Structure Of Pyrimidine Nucleoside 2'-Hydroxylase (Pdn2'H) From Neurospora Crassa In Complex With Thymidine
Organism: Neurospora crassa
Method: X-RAY DIFFRACTION Resolution:1.55 Å Release Date: 2025-02-26 Classification: OXIDOREDUCTASE Ligands: MN, THM, EDO, CL, BU3 |
 |
Crystal Structure Of Human Sirt2 In Complex With The Super-Slow Substrate Tnfn-6
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2024-10-23 Classification: HYDROLASE Ligands: ZN, EDO, BTB, BU3, WU8 |
 |
Crystal Structure Of Human Sirt2 In Complex With The Super-Slow Substrate Tnfn-6 And Nad+
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.55 Å Release Date: 2024-10-23 Classification: HYDROLASE Ligands: ZN, EDO, BTB, DMS, BU3, PGE, NAD, WU8 |
 |
Crystal Structure Of Human Sirt2 In Complex With The Peptide-Based Pseudo-Inhibitor Tnfn-4.1
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.55 Å Release Date: 2024-10-09 Classification: HYDROLASE Ligands: ZN, BU3, EDO, WH8, WGN |