Search Count: 712
 |
Pandda Analysis - Crystal Structure Of Trypanosoma Brucei Trypanothione Reductase In Complex With Z943693514
Organism: Trypanosoma brucei
Method: X-RAY DIFFRACTION Release Date: 2025-11-12 Classification: OXIDOREDUCTASE Ligands: FAD, IMD, PEG, DMS, BR, NA, GT4 |
 |
Pandda Analysis - Crystal Structure Of Trypanosoma Brucei Trypanothione Reductase In Complex With Z2856434836
Organism: Trypanosoma brucei
Method: X-RAY DIFFRACTION Release Date: 2025-11-12 Classification: OXIDOREDUCTASE Ligands: FAD, IMD, PEG, BR, LPZ |
 |
Pandda Analysis - Crystal Structure Of Trypanosoma Brucei Trypanothione Reductase In Complex With Z2204875953
Organism: Trypanosoma brucei
Method: X-RAY DIFFRACTION Release Date: 2025-11-12 Classification: OXIDOREDUCTASE Ligands: FAD, IMD, PEG, Q7L, BR |
 |
Pandda Analysis - Crystal Structure Of Trypanosoma Brucei Trypanothione Reductase In Complex With Z2856434890
Organism: Trypanosoma brucei
Method: X-RAY DIFFRACTION Release Date: 2025-11-12 Classification: OXIDOREDUCTASE Ligands: FAD, IMD, PEG, NZD, BR |
 |
Pandda Analysis - Crystal Structure Of Trypanosoma Brucei Trypanothione Reductase In Complex With Z32399802
Organism: Trypanosoma brucei
Method: X-RAY DIFFRACTION Release Date: 2025-11-12 Classification: OXIDOREDUCTASE Ligands: FAD, IMD, PEG, BR, A1I3U |
 |
Pandda Analysis - Crystal Structure Of Trypanosoma Brucei Trypanothione Reductase In Complex With Z2017861827
Organism: Trypanosoma brucei
Method: X-RAY DIFFRACTION Release Date: 2025-11-12 Classification: OXIDOREDUCTASE Ligands: FAD, IMD, PEG, A1BE9, BR |
 |
Pandda Analysis - Crystal Structure Of Trypanosoma Brucei Trypanothione Reductase In Complex With Z319545618
Organism: Trypanosoma brucei
Method: X-RAY DIFFRACTION Release Date: 2025-11-12 Classification: OXIDOREDUCTASE Ligands: FAD, IMD, PEG, RXS, BR, NA |
 |
Pandda Analysis - Crystal Structure Of Trypanosoma Brucei Trypanothione Reductase In Complex With Z436190540
Organism: Trypanosoma brucei
Method: X-RAY DIFFRACTION Release Date: 2025-11-12 Classification: OXIDOREDUCTASE Ligands: FAD, IMD, PEG, A1I3V, BR, NA |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-11-12 Classification: TRANSFERASE/Inhibitor Ligands: A1CNU, BR, CL, NA, TME |
 |
Organism: Aspergillus niger
Method: X-RAY DIFFRACTION Release Date: 2025-11-05 Classification: OXIDOREDUCTASE Ligands: EDO, MAN, NAG, FAD, CL, NA, BA, BR |
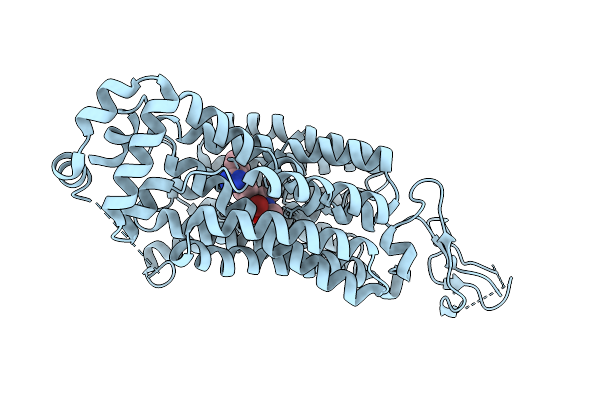 |
Cryo-Em Structure Of Human Oat1 In Complex With Olmesartan And Bromide Ion.
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-11-05 Classification: MEMBRANE PROTEIN Ligands: BR, OLM |
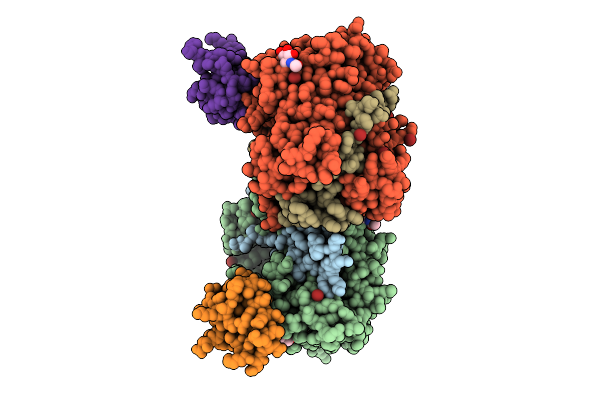 |
Structure Of Native Leukocyte Myeloperoxidase In Complex With A Truncated Version Of The Staphylococcal Peroxidase Inhibitor Spin And Bromide At Ph 7.5
Organism: Staphylococcus aureus, Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-10-15 Classification: OXIDOREDUCTASE Ligands: HEM, BR, NAG, CA |
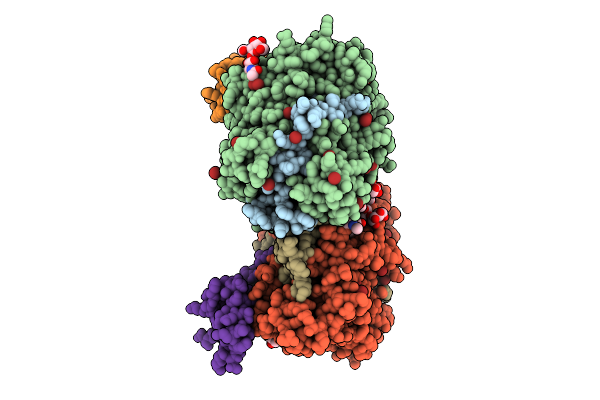 |
Structure Of Native Leukocyte Myeloperoxidase In Complex With The Staphylococcal Peroxidase Inhibitor Spin And Bromide At Ph 5.5
Organism: Staphylococcus aureus, Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-10-15 Classification: OXIDOREDUCTASE Ligands: NAG, BR, CA, HEM |
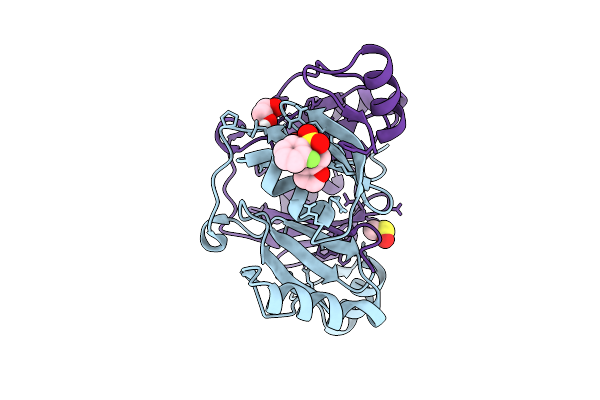 |
Fragment Screening Of Fosakp, Cryo Structure In Complex With Fragment F2X-Entry D07
Organism: Klebsiella pneumoniae
Method: X-RAY DIFFRACTION Release Date: 2025-10-15 Classification: TRANSFERASE Ligands: SYJ, EDO, MN, BR, DMS |
 |
Crystal Structure Of Dna Integrity Scanning Protein Disa From Mycobacterium Tuberculosis In Complex With Cyclic Di-Amp And Bromide
Organism: Mycobacterium tuberculosis h37rv
Method: X-RAY DIFFRACTION Release Date: 2025-09-24 Classification: TRANSFERASE Ligands: 2BA, BR, CL |
 |
Usp7 Covalently Bound To N-(6-Fluoro-3-Nitropyridin-2-Yl)-5-(1-Methyl-1H-Pyrazol-4-Yl)Isoquinolin-3-Amine (Gcl36, 7A) With Partial Occupancy
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-09-10 Classification: HYDROLASE Ligands: A1I71, EDO, PEG, BR |
 |
Pandda Analysis Group Deposition -- Crystal Structure Of Sars-Cov-2 Nsp3 Macrodomain In Complex With Avi-0000372
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Release Date: 2025-08-20 Classification: VIRAL PROTEIN Ligands: DMS, A1A7P, BR |
 |
Organism: Oryctolagus cuniculus
Method: X-RAY DIFFRACTION Release Date: 2025-08-06 Classification: HYDROLASE Ligands: TB, CA, OCH, BR |
 |
Crystal Structure Of A Galactose Oxidase From Pseudoarthrobacter Siccitolerans
Organism: Pseudarthrobacter siccitolerans
Method: X-RAY DIFFRACTION Release Date: 2025-07-23 Classification: OXIDOREDUCTASE Ligands: GOL, PG4, CA, CL, BR |
 |
X-Ray Crystal Structure Of Sars-Cov-2 Main Protease Quadruple Mutants In Complex With Nirmatrelvir
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Release Date: 2025-07-09 Classification: HYDROLASE/INHIBITOR Ligands: 4WI, EDO, BR, SO4 |

