Search Count: 21
 |
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Release Date: 2025-11-26 Classification: DE NOVO PROTEIN Ligands: I77, BNZ, I6W |
 |
Uic-1 Soaked In Equimolar Benzene, Toluene, Isopropylbenzene, And Ortho-Xylene
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Release Date: 2025-11-26 Classification: DE NOVO PROTEIN Ligands: I77, I6W, PYJ, BNZ, IPB, OXE |
 |
Organism: Trypanosoma brucei brucei
Method: X-RAY DIFFRACTION Resolution:2.75 Å Release Date: 2019-12-25 Classification: TRANSFERASE Ligands: BNZ, GOL, AMP |
 |
Organism: Enterobacteria phage t4
Method: X-RAY DIFFRACTION Resolution:1.41 Å Release Date: 2016-09-21 Classification: HYDROLASE Ligands: BNZ |
 |
Organism: Enterobacteria phage t4
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2015-04-01 Classification: HYDROLASE Ligands: BNZ, EPE |
 |
Organism: Enterobacteria phage t4
Method: X-RAY DIFFRACTION Resolution:1.67 Å Release Date: 2013-03-27 Classification: HYDROLASE Ligands: BME, BNZ, SO4, ACT, HED |
 |
Structural Determinants Of The Beta-Selectivity Of A Bacterial Aminotransferase
Organism: Mesorhizobium sp. luk
Method: X-RAY DIFFRACTION Resolution:1.69 Å Release Date: 2012-05-30 Classification: TRANSFERASE Ligands: EDO, PLP, SFE, BNZ, GOL |
 |
New Azaborine Compounds Bind To The T4 Lysozyme L99A Cavity - Benzene As Control
Organism: Enterobacteria phage t4
Method: X-RAY DIFFRACTION Resolution:1.25 Å Release Date: 2009-10-13 Classification: HYDROLASE Ligands: HED, PO4, BNZ |
 |
T4 Lysozyme M102E/L99A Mutant With Buried Charge In Apolar Cavity--Benzene Binding
Organism: Enterobacteria phage t4
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2009-08-25 Classification: HYDROLASE Ligands: BNZ, BME, CO3, EPE, SO4 |
 |
Organism: Bacteriophage t4
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2009-01-27 Classification: HYDROLASE Ligands: PO4, CL, HED, BNZ |
 |
Organism: Sars coronavirus
Method: X-RAY DIFFRACTION Resolution:1.86 Å Release Date: 2007-12-25 Classification: HYDROLASE Ligands: HG, BNZ |
 |
Urate Oxidase From Aspergillus Flavus Complexed With 5-Amino 6-Nitro Uracil
Organism: Aspergillus flavus
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2005-03-22 Classification: OXIDOREDUCTASE Ligands: UNC, BNZ |
 |
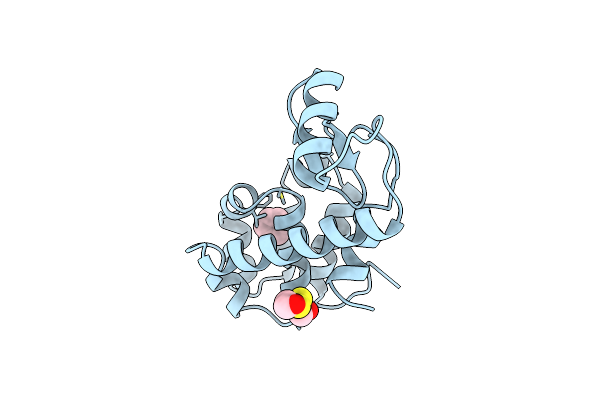
Organism: Streptomyces fradiae
Method: X-RAY DIFFRACTION Resolution:0.95 Å Release Date: 1999-03-23 Classification: ANTIBIOTIC Ligands: BNZ |
 |
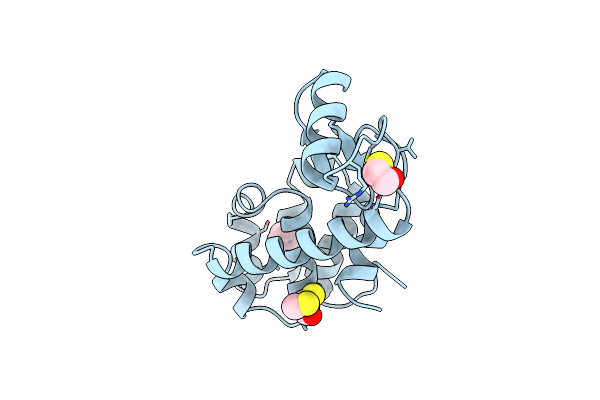
Generating Ligand Binding Sites In T4 Lysozyme Using Deficiency-Creating Substitutions
Organism: Enterobacteria phage t4
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 1998-03-18 Classification: HYDROLASE Ligands: CL, BME, BNZ |
 |
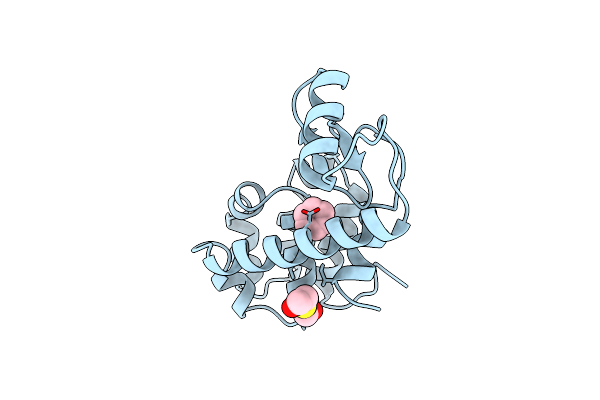
Generating Ligand Binding Sites In T4 Lysozyme Using Deficiency-Creating Substitutions
Organism: Enterobacteria phage t4
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 1998-03-18 Classification: HYDROLASE Ligands: CL, BME, BNZ |
 |
Generating Ligand Binding Sites In T4 Lysozyme Using Deficiency-Creating Substitutions
Organism: Enterobacteria phage t4
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 1998-03-18 Classification: HYDROLASE Ligands: CL, BME, BNZ |
 |
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 1996-11-08 Classification: LEUCINE ZIPPER Ligands: BNZ |
 |
Specificity Of Ligand Binding In A Buried Non-Polar Cavity Of T4 Lysozyme: Linkage Of Dynamics And Structural Plasticity
Organism: Enterobacteria phage t4
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 1995-07-10 Classification: HYDROLASE (O-GLYCOSYL) Ligands: CL, HED, BNZ |
 |
Organism: Enterobacteria phage t4
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 1993-10-31 Classification: HYDROLASE(O-GLYCOSYL) Ligands: CL, BME, BNZ |
 |
Organism: Enterobacteria phage t4
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 1993-10-31 Classification: HYDROLASE(O-GLYCOSYL) Ligands: CL, BME, BNZ |

