Search Count: 91
All
Selected
 |
Lipidic Cubic Phase Serial Femtosecond Crystallography Structure Of A Photosynthetic Reaction Centre
Organism: Blastochloris viridis
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2022-06-22 Classification: MEMBRANE PROTEIN Ligands: HEC, DGA, HTO, OLC, SO4, BCB, BPB, UQ1, LDA, FE2, MQ7, NS5 |
 |
Lipidic Cubic Phase Serial Femtosecond Crystallography Structure Of A Photosynthetic Reaction Centre
Organism: Blastochloris viridis
Method: X-RAY DIFFRACTION Resolution:2.25 Å Release Date: 2022-06-22 Classification: MEMBRANE PROTEIN Ligands: HEC, DGA, LDA, SO4, BCB, BPB, HTO, UQ2, FE2, MQ9, NS5, OLC |
 |
Ultrafast Structural Response To Charge Redistribution Within A Photosynthetic Reaction Centre - 1 Ps Structure
Organism: Blastochloris viridis
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2020-12-09 Classification: ELECTRON TRANSPORT Ligands: HEC, DGA, SO4, LDA, HTO, BCB, BPB, FE, MQ7, NS5 |
 |
Ultrafast Structural Response To Charge Redistribution Within A Photosynthetic Reaction Centre - 5 Ps (A) Structure
Organism: Blastochloris viridis
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2020-12-09 Classification: ELECTRON TRANSPORT Ligands: HEC, DGA, SO4, LDA, HTO, BCB, BPB, FE, MQ7, NS5 |
 |
Ultrafast Structural Response To Charge Redistribution Within A Photosynthetic Reaction Centre - 300 Ps (A) Structure
Organism: Blastochloris viridis
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2020-12-09 Classification: ELECTRON TRANSPORT Ligands: HEC, DGA, SO4, LDA, HTO, BCB, BPB, FE, MQ7, NS5 |
 |
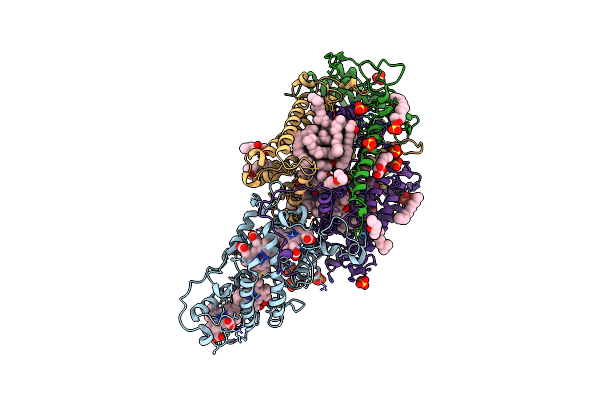
Ultrafast Structural Response To Charge Redistribution Within A Photosynthetic Reaction Centre - 20 Ps Structure
Organism: Blastochloris viridis
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2020-12-09 Classification: ELECTRON TRANSPORT Ligands: HEC, DGA, SO4, LDA, HTO, BCB, BPB, FE, MQ7, NS5 |
 |
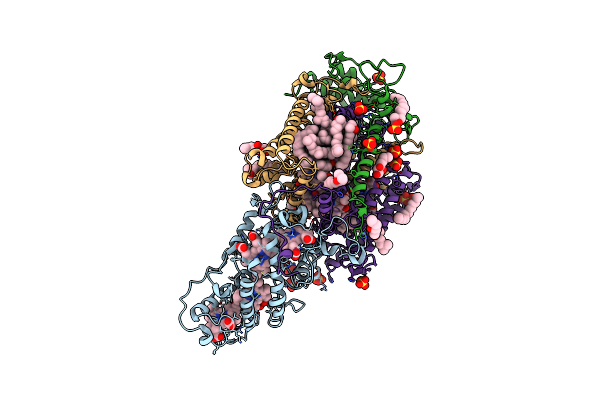
Ultrafast Structural Response To Charge Redistribution Within A Photosynthetic Reaction Centre - 300 Ps (B) Structure
Organism: Blastochloris viridis
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2020-12-09 Classification: ELECTRON TRANSPORT Ligands: HEC, DGA, SO4, LDA, HTO, BCB, BPB, FE, MQ7, NS5 |
 |
Ultrafast Structural Response To Charge Redistribution Within A Photosynthetic Reaction Centre - 8 Us Structure
Organism: Blastochloris viridis
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2020-12-09 Classification: ELECTRON TRANSPORT Ligands: HEC, DGA, SO4, LDA, HTO, BCB, BPB, FE, MQ7, NS5 |
 |
Ultrafast Structural Response To Charge Redistribution Within A Photosynthetic Reaction Centre - 5 Ps (B) Structure
Organism: Blastochloris viridis
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2020-12-09 Classification: ELECTRON TRANSPORT Ligands: HEC, DGA, SO4, LDA, HTO, BCB, BPB, FE, MQ7, NS5 |
 |
Crystal Structure Of The Homospermidine Synthase (Hss) Variant W229E From Blastochloris Viridis In Complex With Nad
Organism: Blastochloris viridis
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2020-08-26 Classification: TRANSFERASE Ligands: NAD |
 |
Crystal Structure Of The Homospermidine Synthase (Hss) Variant N135F From Blastochloris Viridis In Complex With Nad
Organism: Blastochloris viridis
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2020-07-15 Classification: TRANSFERASE Ligands: NAD |
 |
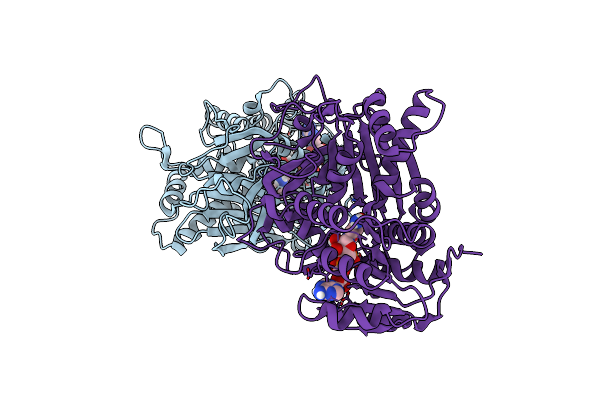
Crystal Structure Of The Homospermidine Synthase (Hss) Variant E117Q From Blastochloris Viridis In Complex With Nad
Organism: Blastochloris viridis
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2020-07-15 Classification: TRANSFERASE Ligands: NAD, SO4, 1PS |
 |
Crystal Structure Of The Homospermidine Synthase (Hss) Variant W229A From Blastochloris Viridis In Complex With Nad And Put
Organism: Blastochloris viridis
Method: X-RAY DIFFRACTION Resolution:1.87 Å Release Date: 2020-07-15 Classification: TRANSFERASE Ligands: NAD, PUT |
 |
Crystal Structure Of The Homospermidine Synthase (Hss) Variant E210Q From Blastochloris Viridis In Complex With Nad
Organism: Blastochloris viridis
Method: X-RAY DIFFRACTION Resolution:1.72 Å Release Date: 2020-07-08 Classification: TRANSFERASE Ligands: SO4, AG2, NAD |
 |
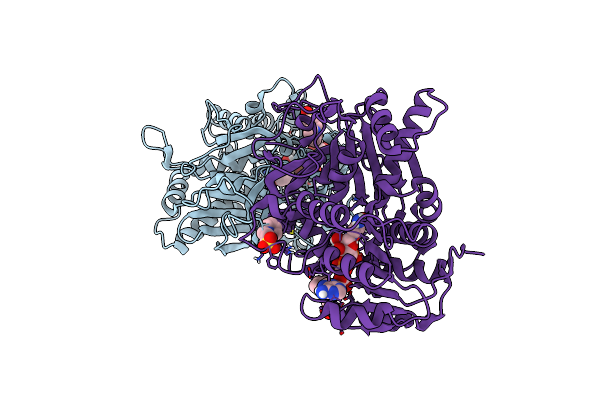
Crystal Structure Of The Homospermidine Synthase (Hss) Variant E210A From Blastochloris Viridis In Complex With Nad
Organism: Blastochloris viridis
Method: X-RAY DIFFRACTION Resolution:1.69 Å Release Date: 2020-07-08 Classification: TRANSFERASE Ligands: AG2, SO4, NAD |
 |
Crystal Structure Of The Homospermidine Synthase (Hss) Variant W229F From Blastochloris Viridis In Complex With Nad
Organism: Blastochloris viridis
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2020-07-08 Classification: TRANSFERASE Ligands: NAD, 1PS |
 |
Organism: Blastochloris viridis
Method: ELECTRON MICROSCOPY Resolution:2.87 Å Release Date: 2018-04-11 Classification: PHOTOSYNTHESIS Ligands: HEC, BCB, BPB, UQ9, FE, SO4, LDA, MQ9, NS5, NS0 |
 |
From Macrocrystals To Microcrystals: A Strategy For Membrane Protein Serial Crystallography
Organism: Blastochloris viridis
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2017-08-16 Classification: MEMBRANE PROTEIN Ligands: HEC, DGA, SO4, LDA, HTO, BCB, BPB, FE2, MQ7, NS5 |
 |
From Macrocrystals To Microcrystals: A Strategy For Membrane Protein Serial Crystallography
Organism: Blastochloris viridis
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2017-08-16 Classification: MEMBRANE PROTEIN Ligands: HEC, DGA, SO4, LDA, HTO, BCB, BPB, FE2, MQ7, NS5 |
 |
From Macrocrystals To Microcrystals: A Strategy For Membrane Protein Serial Crystallography
Organism: Blastochloris viridis
Method: X-RAY DIFFRACTION Resolution:3.30 Å Release Date: 2017-08-16 Classification: MEMBRANE PROTEIN Ligands: HEC, DGA, SO4, FME, LDA, HTO, BCB, BPB, FE2, MQ7, NS5 |

