Search Count: 16
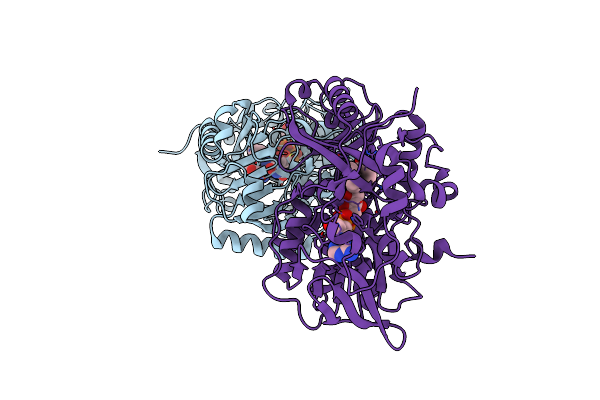 |
Structure Of L-Amino Acid Deaminase From Proteus Myxofaciens In Complex With Anthranilate
Organism: Proteus myxofaciens
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2016-04-06 Classification: HYDROLASE Ligands: FAD, BE2 |
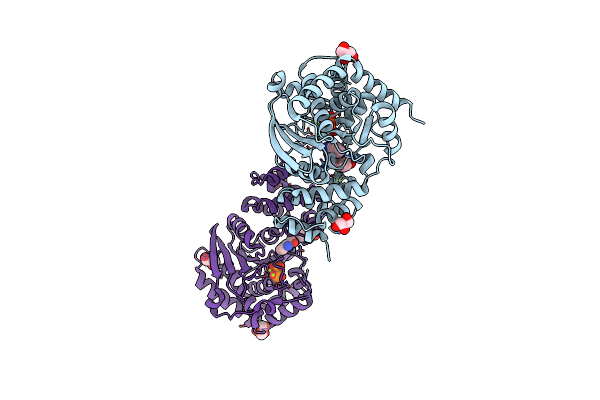 |
Anthranilate Phosphoribosyltransferase Variant R194A From Mycobacterium Tuberculosis With Pyrophosphate, Mg2+ And Anthranilate Bound
Organism: Mycobacterium tuberculosis
Method: X-RAY DIFFRACTION Resolution:1.77 Å Release Date: 2015-11-25 Classification: TRANSFERASE Ligands: POP, MG, BE2, GOL |
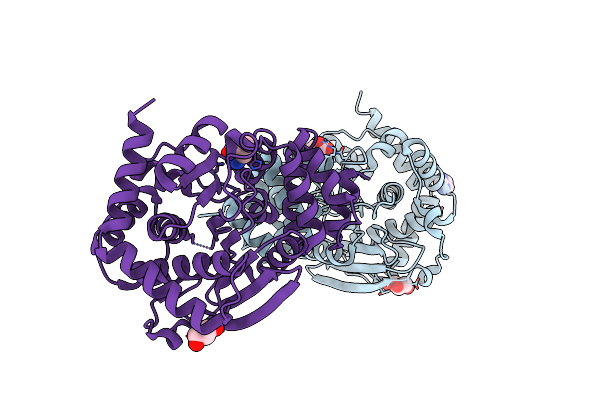 |
Anthranilate Phosphoribosyltransferase Variant R193A From Mycobacterium Tuberculosis With Anthranilate Bound
Organism: Mycobacterium tuberculosis
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2015-09-23 Classification: TRANSFERASE Ligands: BE2, IMD, GOL |
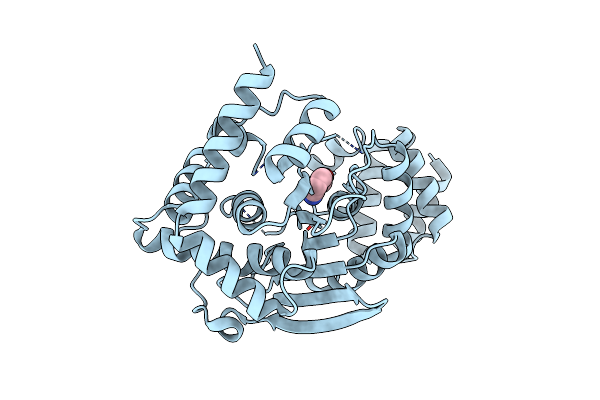 |
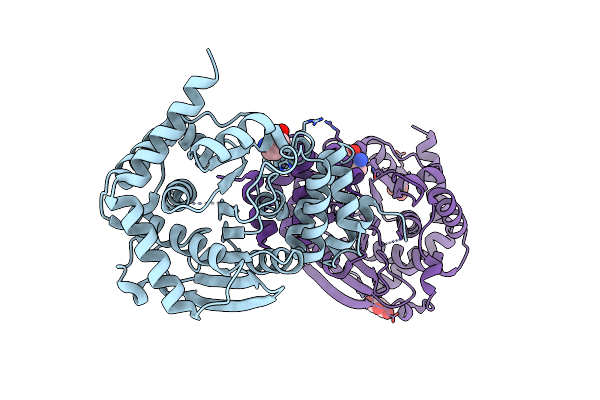
Anthranilate Bound At Active Site Of Anthranilate Phosphoribosyl Transferase From Acinetobacter (Anprt; Trpd)
Organism: Acinetobacter baylyi (strain atcc 33305 / bd413 / adp1)
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2015-09-02 Classification: TRANSFERASE Ligands: BE2 |
 |
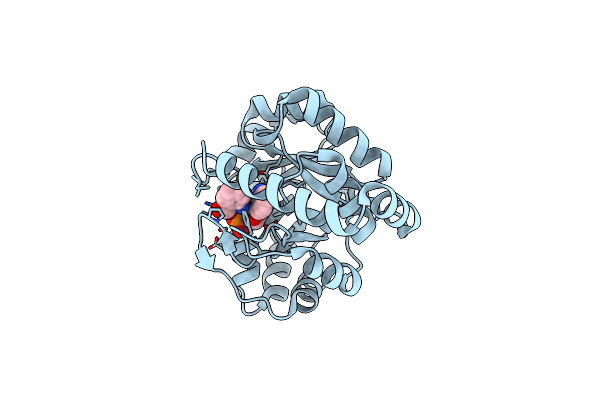
Anthranilate Phosphoribosyl Transferase From Mycobacterium Tuberculosis In Complex With Anthranilate
Organism: Mycobacterium tuberculosis
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2014-04-23 Classification: TRANSFERASE Ligands: BE2, GOL, PO4 |
 |
Crystal Structure Of Mycobacterium Tuberculosis Indole Glycerol Phosphate Synthase (Igps) In Complex With Indole Glycerol Phosphate (Igp) Amd Anthranilate
Organism: Mycobacterium tuberculosis
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2012-08-08 Classification: TRANSFERASE Ligands: IGP, BE2 |
 |
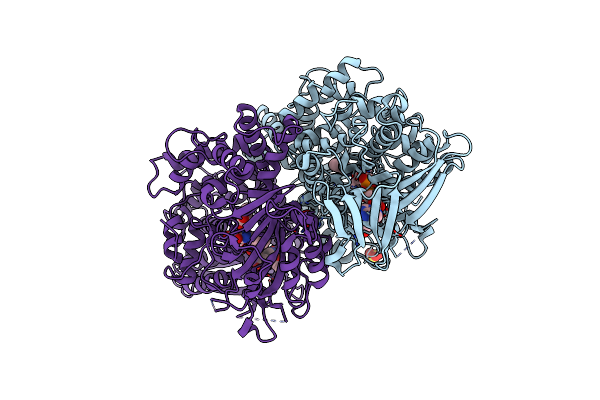
Crystal Structure Of Pseudomonas Aeruginosa Pqsd C112A Mutant In Complex With Anthranilic Acid
Organism: Pseudomonas aeruginosa pao1
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2009-09-15 Classification: TRANSFERASE Ligands: BE2 |
 |
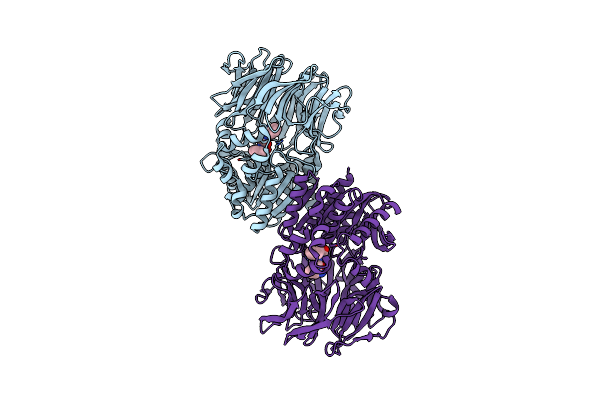
Organism: Pseudomonas sp. p-501
Method: X-RAY DIFFRACTION Resolution:1.35 Å Release Date: 2008-04-15 Classification: OXIDOREDUCTASE Ligands: SO4, FAD, BE2, GOL |
 |
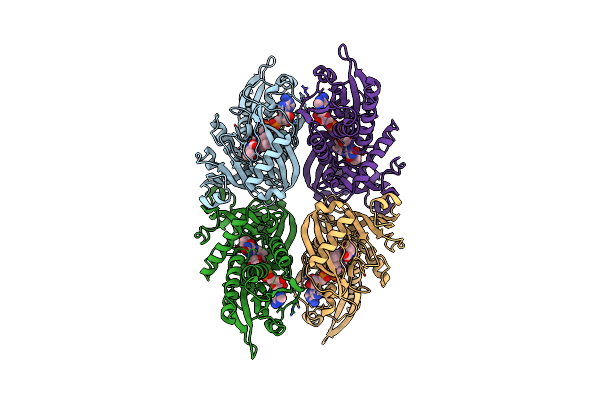
Organism: Aeropyrum pernix
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2007-05-15 Classification: HYDROLASE Ligands: BE2, GLY, PHE |
 |
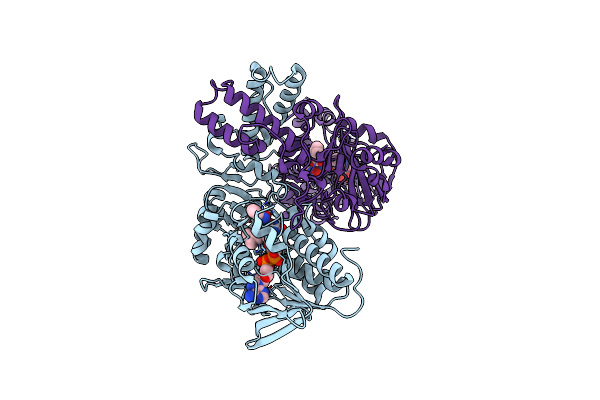
Crystal Structure Of Human D-Amino Acid Oxidase In Complex With O-Aminobenzoate
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2007-03-06 Classification: OXIDOREDUCTASE Ligands: FAD, BE2 |
 |
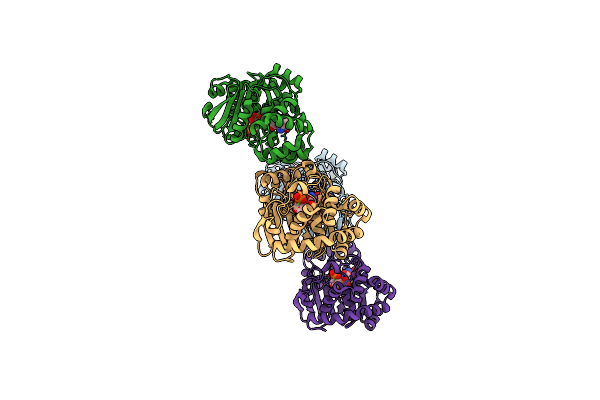
The Structure Of L-Amino Acid Oxidase From Rhodococcus Opacus In Complex With O-Aminobenzoate
Organism: Rhodococcus opacus
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2007-01-30 Classification: OXIDOREDUCTASE Ligands: FAD, BE2 |
 |
Anthranilate Phosphoribosyltransferase In Complex With Prpp, Anthranilate And Magnesium
Organism: Sulfolobus solfataricus
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2006-05-23 Classification: TRANSFERASE Ligands: MG, PRP, BE2 |
 |
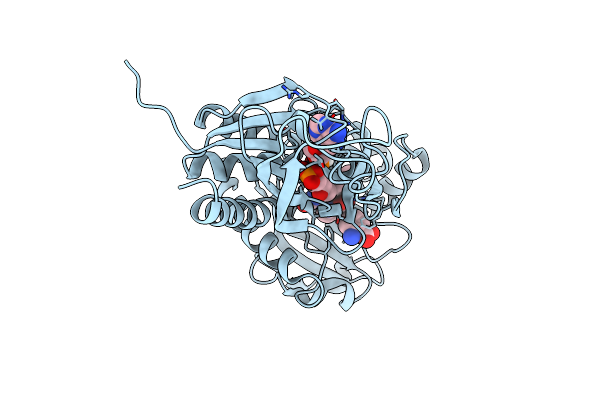
Anthranilate Phosphoribosyl-Transferase (Trpd) From S. Solfataricus In Complex With Anthranilate
Organism: Sulfolobus solfataricus
Method: X-RAY DIFFRACTION Resolution:2.43 Å Release Date: 2006-05-23 Classification: TRANSFERASE Ligands: BE2 |
 |
Crystal Structure Of D-Amino Acid Oxidase In Complex With Two Anthranylate Molecules
Organism: Rhodosporidium toruloides
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2002-02-27 Classification: OXIDOREDUCTASE Ligands: FAD, BE2 |
 |
Crystal Structure Of L-Amino Acid Oxidase From Calloselasma Rhodostoma, Complexed With Three Molecules Of O-Aminobenzoate.
Organism: Calloselasma rhodostoma
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2000-08-24 Classification: OXIDOREDUCTASE Ligands: NAG, BE2, FAD |
 |
Organism: Sus scrofa
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 1997-11-12 Classification: OXIDOREDUCTASE Ligands: FAD, BE2 |

