Search Count: 30
 |
Organism: Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2025-06-11 Classification: PROTEIN FIBRIL Ligands: BDP |
 |
Organism: Roseburia hominis
Method: X-RAY DIFFRACTION Resolution:2.90 Å Release Date: 2024-03-20 Classification: HYDROLASE Ligands: GOL, BDP, FMN, ZG5 |
 |
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.37 Å Release Date: 2022-11-16 Classification: HYDROLASE Ligands: ZN, SO4, C3G, BDP |
 |
Organism: Unidentified
Method: X-RAY DIFFRACTION Resolution:2.15 Å Release Date: 2022-10-12 Classification: HYDROLASE Ligands: BDP, ACT, ZN, SO4 |
 |
Crystal Structure Of Thermotoga Maritima Alpha-Glucuronidase (Tm0752) In Complex With Nadh And D-Glucuronic Acid
Organism: Thermotoga maritima (strain atcc 43589 / msb8 / dsm 3109 / jcm 10099)
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2021-09-01 Classification: HYDROLASE Ligands: NAI, BDP, IPA |
 |
Organism: Ruminococcus gnavus
Method: X-RAY DIFFRACTION Resolution:1.58 Å Release Date: 2020-06-10 Classification: HYDROLASE Ligands: BDP |
 |
The Glucuronoyl Esterase Otce15A S267A Variant From Opitutus Terrae In Complex With D-Glucuronate
Organism: Opitutus terrae pb90-1
Method: X-RAY DIFFRACTION Resolution:1.12 Å Release Date: 2019-11-27 Classification: HYDROLASE Ligands: MG, EDO, GCU, BDP, PEG |
 |
The Glucuronoyl Esterase Otce15A H408A Variant From Opitutus Terrae In Complex With, And Covalently Linked To, D-Glucuronate
Organism: Opitutus terrae pb90-1
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2019-11-27 Classification: HYDROLASE Ligands: BDP, FMT |
 |
Structure Of Bifidobacterium Dentium Beta-Glucuronidase Complexed With Coumarin-3-O-Glucuronide
Organism: Bifidobacterium dentium atcc 27679
Method: X-RAY DIFFRACTION Resolution:1.45 Å Release Date: 2018-12-12 Classification: HYDROLASE Ligands: BDP, SE2 |
 |
Organism: Eubacterium eligens (strain atcc 27750 / vpi c15-48)
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2018-07-25 Classification: HYDROLASE Ligands: BDP |
 |
Organism: Bacteroides thetaiotaomicron (strain atcc 29148 / dsm 2079 / nctc 10582 / e50 / vpi-5482)
Method: X-RAY DIFFRACTION Resolution:1.39 Å Release Date: 2017-04-26 Classification: HYDROLASE Ligands: BDP |
 |
Crystal Structure Of A Trap Periplasmic Solute Binding Protein From Pseudomonas Putida F1 (Pput_1203), Target Efi-500184, With Bound D-Glucuronate
Organism: Pseudomonas putida nd6
Method: X-RAY DIFFRACTION Resolution:1.40 Å Release Date: 2015-01-28 Classification: TRANSPORT PROTEIN Ligands: BDP, CXS, SO4 |
 |
Crystal Structure Of A Trap Periplasmic Solute Binding Protein From Rhodobacter Sphaeroides (Rsph17029_2138, Target Efi-510205) With Bound Glucuronate
Organism: Rhodobacter sphaeroides (strain atcc 17029 / ath 2.4.9)
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2014-12-24 Classification: SOLUTE-BINDING PROTEIN Ligands: BDP, PO4 |
 |
Crystal Structure Of A Trap Periplasmic Solute Binding Protein From Citrobacter Koseri (Cko_04899, Target Efi-510094) With Bound D-Glucuronate
Organism: Citrobacter koseri
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2014-12-10 Classification: SOLUTE BINDING PROTEIN Ligands: BDP, CL, MG |
 |
Organism: Paenibacillus barcinonensis
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2014-10-08 Classification: SUGAR BINDING PROTEIN Ligands: GCU, CA, BDP |
 |
Crystal Structure Of A Trap Periplasmic Solute Binding Protein From Chromohalobacter Salexigens Dsm 3043 (Csal_2479), Target Efi-510085, With Bound Glucuronate, Spg P6122
Organism: Chromohalobacter salexigens
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2014-04-02 Classification: SOLUTE-BINDING PROTEIN Ligands: CL, BDP |
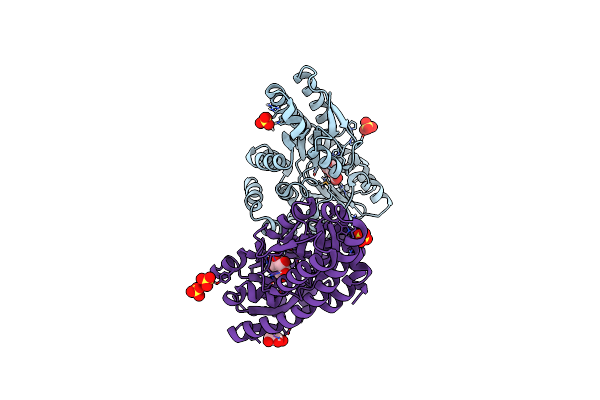 |
Crystal Structure Of A Trap Periplasmic Solute Binding Protein From Chromohalobacter Salexigens Dsm 3043 (Csal_2479), Target Efi-510085, With Bound D-Glucuronate, Spg I213
Organism: Chromohalobacter salexigens
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2014-03-19 Classification: TRANSPORT PROTEIN Ligands: BDP, SO4, GOL |
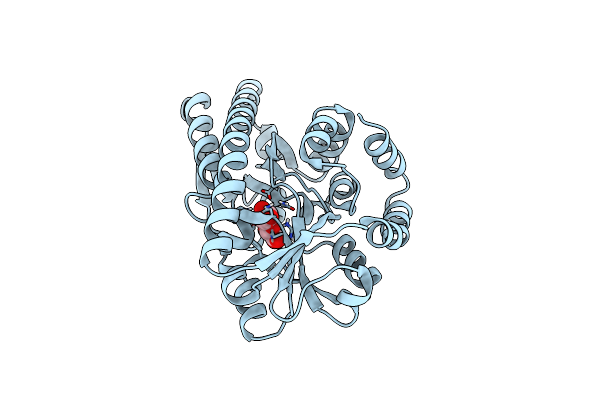 |
Crystal Structure Of A Trap Periplasmic Solute Binding Protein From Roseobacter Denitrificans, Target Efi-510230, With Bound Beta-D-Glucuronate
Organism: Roseobacter denitrificans
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2014-01-22 Classification: SOLUTE-BINDING PROTEIN Ligands: CL, BDP |
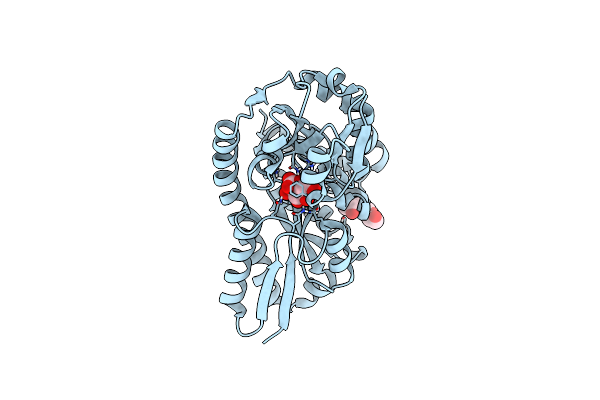 |
Crystal Structure Of A Trap Periplasmic Solute Binding Protein From Anaerococcus Prevotii Dsm 20548 (Apre_1383), Target Efi-510023, With Bound Alpha/Beta D-Glucuronate
Organism: Anaerococcus prevoti
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2013-11-13 Classification: TRANSPORT PROTEIN Ligands: GCU, BDP, CL, PG4 |
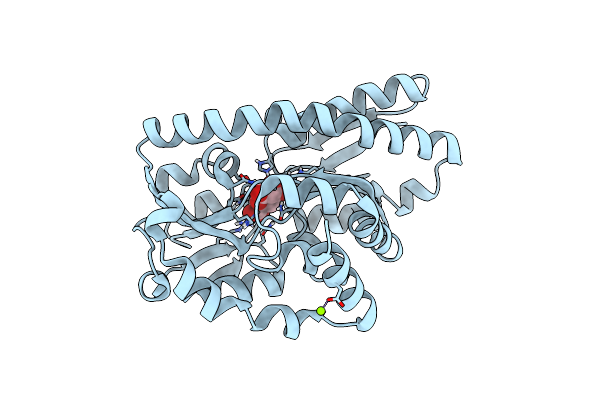 |
Crystal Structure Of A Trap Periplasmic Solute Binding Protein From Burkholderia Ambifaria (Bam_6123), Target Efi-510059, With Bound Beta-D-Glucuronate
Organism: Burkholderia ambifaria
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2013-10-16 Classification: TRANSPORT PROTEIN Ligands: MG, BDP |

