Search Count: 13
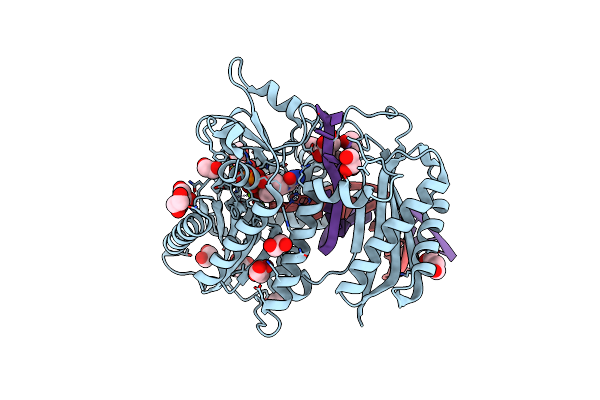 |
Organism: Saccharomyces cerevisiae (strain atcc 204508 / s288c), Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2017-10-25 Classification: TRANSFERASE/DNA Ligands: MG, BAP, EDO, DCP, GOL, PEG |
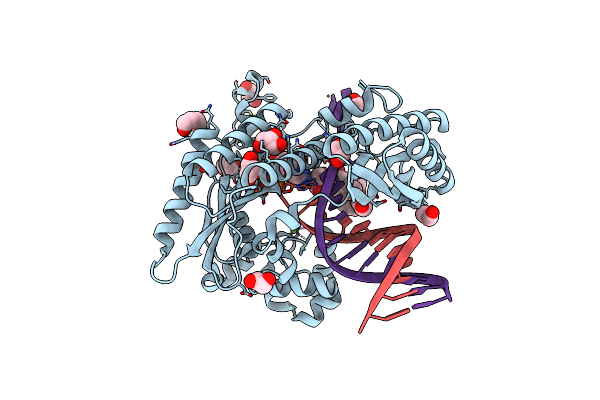 |
Organism: Saccharomyces cerevisiae (strain atcc 204508 / s288c), Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.92 Å Release Date: 2017-10-25 Classification: TRANSFERASE/DNA Ligands: MG, BAP, DCP, GOL, EDO, PEG |
 |
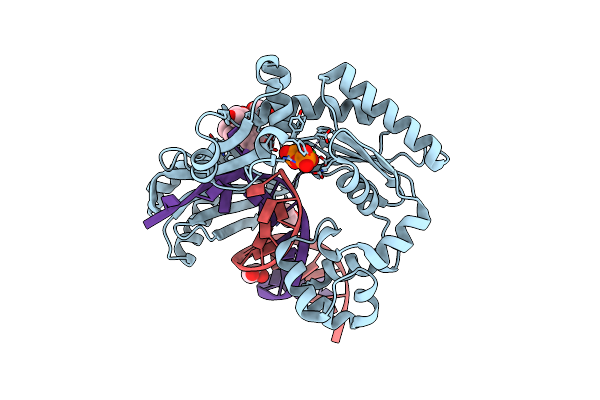
Bypass Of Major Benzopyrene-Dg Adduct By Y-Family Dna Polymerase With Unique Structural Gap
Organism: Sulfolobus solfataricus
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2007-09-11 Classification: TRANSFERASE/DNA Ligands: PO4, CA, ATP, GOL, EDO, BAP |
 |
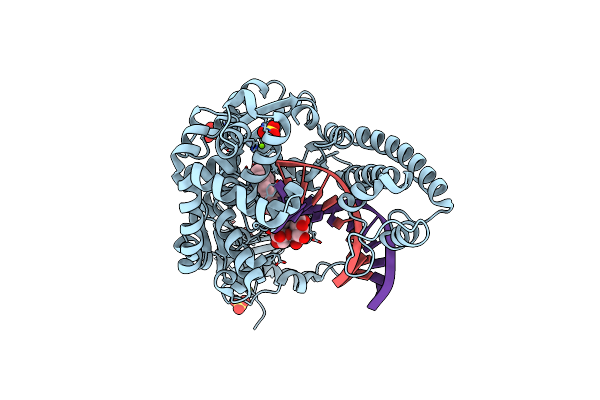
Bypass Of Major Benzopyrene-Dg Adduct By Y-Family Dna Polymerase With Unique Structural Gap
Organism: Sulfolobus solfataricus
Method: X-RAY DIFFRACTION Resolution:2.25 Å Release Date: 2007-09-11 Classification: TRANSFERASE/DNA Ligands: BAP, EDO, GOL, CA, POP |
 |
Methylation Of Cytosine At C5 In A Cpg Sequence Context Causes A Conformational Switch Of A Benzo[A]Pyrene Diol Epoxide-N2-Guanine Adduct In Dna From A Minor Groove Alignment To Intercalation With Base Displacement
|
 |
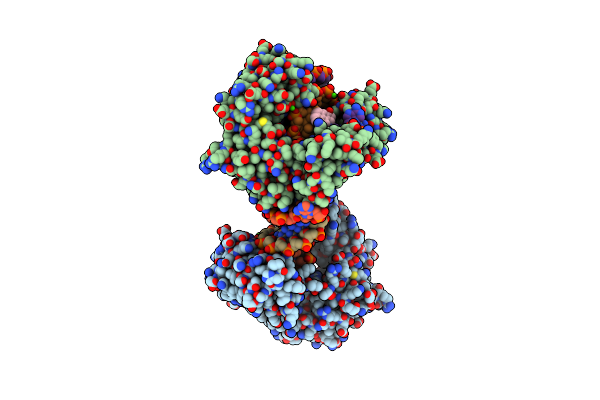
Structure Of A High-Fidelity Polymerase Bound To A Benzo[A]Pyrene Adduct That Blocks Replication
Organism: Geobacillus stearothermophilus
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2004-12-14 Classification: TRANSFERASE/DNA Ligands: BAP, SO4, MG |
 |
Crystal Structure Of A Benzo[A]Pyrene Diol Epoxide Adduct In A Ternary Complex With A Dna Polymerase
Organism: Sulfolobus solfataricus
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2004-03-30 Classification: TRANSFERASE/DNA Ligands: BAP, CA, MG, DTP |
 |
 |
Minor Conformer Of A Benzo[A]Pyrene Diol Epoxide Adduct Of Da In Duplex Dna
Organism: Synthetic construct
Method: SOLUTION NMR Release Date: 1998-08-19 Classification: DNA Ligands: BAP |
 |
Solution Conformation Of The (-)-Trans-Anti-[Bp]Dg Adduct Opposite A Deletion Site In Dna Duplex D(Ccatc-[Bp]G-Ctacc)D(Ggtag--Gatgg), Nmr, 6 Structures
|
 |
Structural Alignment Of The (+)-Trans-Anti-[Bp]Dg Adduct Positioned Opposite Dc At A Dna Template-Primer Junction, Nmr, 6 Structures
|
 |
Solution Nmr Structure Of The [Bp]Da Adduct Opposite Dt In A Dna Duplex, 6 Structures
|
 |

