Search Count: 232
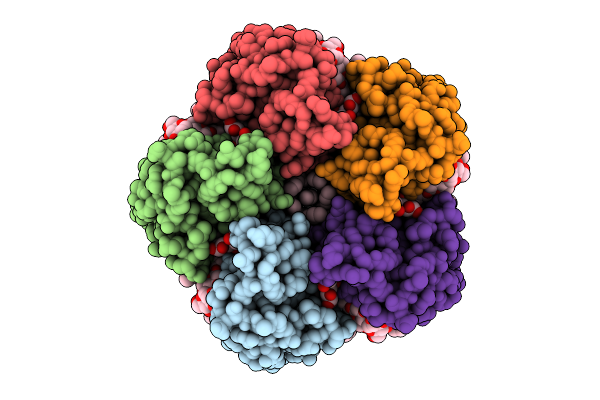 |
Cryo-Em Structure Of The Light-Driven Sodium Pump Ernar In The Pentameric Form
Organism: Erythrobacter
Method: ELECTRON MICROSCOPY Resolution:2.10 Å Release Date: 2025-12-17 Classification: MEMBRANE PROTEIN Ligands: LFA, LMT, RET |
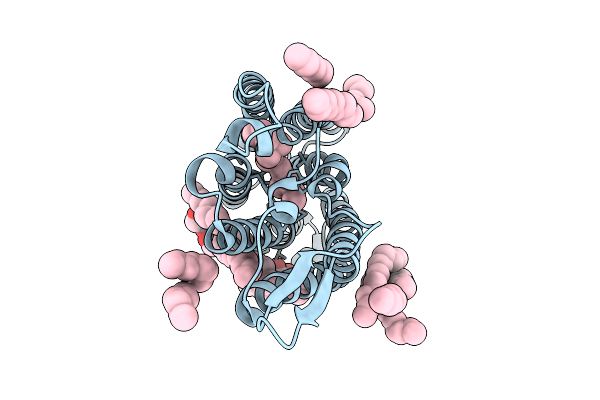 |
Cryo-Em Structure Of The Light-Driven Sodium Pump Ernar In The Monomeric Form In The K2 State
Organism: Erythrobacter
Method: ELECTRON MICROSCOPY Resolution:2.80 Å Release Date: 2025-12-17 Classification: MEMBRANE PROTEIN Ligands: LFA, LMT, RET |
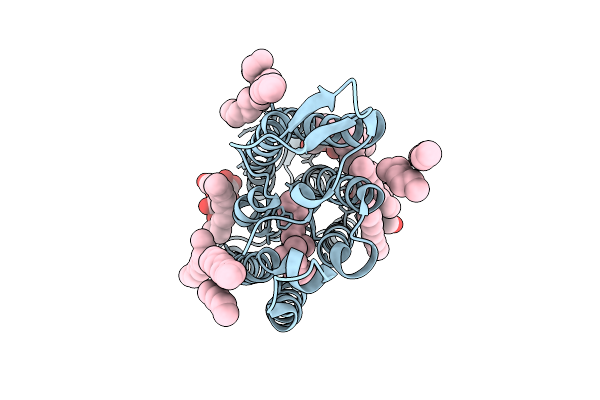 |
Cryo-Em Structure Of The Light-Driven Sodium Pump Ernar In The Monomeric Form In The O2 State
Organism: Erythrobacter
Method: ELECTRON MICROSCOPY Resolution:3.30 Å Release Date: 2025-12-17 Classification: MEMBRANE PROTEIN Ligands: LFA, LMT, RET |
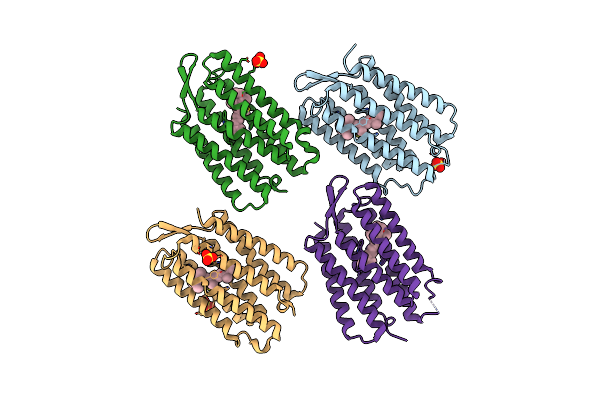 |
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.76 Å Release Date: 2025-06-04 Classification: DE NOVO PROTEIN Ligands: RET, SO4 |
 |
Organism: Haloquadratum walsbyi dsm 16790
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2025-05-21 Classification: MEMBRANE PROTEIN Ligands: RET, MG |
 |
Anabaena Sensory Rhodopsin Structure Determination From Paramagnetic Relaxation Enhancement And Nmr Restraints
Organism: Nostoc sp.
Method: SOLID-STATE NMR Release Date: 2025-04-16 Classification: MEMBRANE PROTEIN Ligands: RET |
 |
Crystal Structure Of Marine Actinobacteria Clade Rhodopsin (Mar) In The Ground State
Organism: Candidatus actinomarina minuta, Marine actinobacteria clade
Method: X-RAY DIFFRACTION Resolution:1.25 Å Release Date: 2025-04-02 Classification: MEMBRANE PROTEIN Ligands: OLC, LFA, GOL, RET, PO4 |
 |
Crystal Structure Of Marine Actinobacteria Clade Rhodopsin (Mar) In The P596 State
Organism: Candidatus actinomarina minuta, Marine actinobacteria clade
Method: X-RAY DIFFRACTION Resolution:1.41 Å Release Date: 2025-04-02 Classification: MEMBRANE PROTEIN Ligands: OLC, LFA, GOL, RET, PO4 |
 |
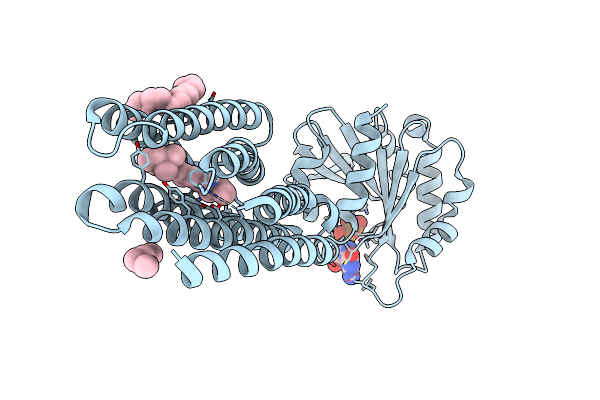
Crystal Structure Of Marine Actinobacteria Clade Rhodopsin (Mar) - Human Gtpase Arf1 (L8K,Q71L) Chimera; Ground State
Organism: Candidatus actinomarina minuta, Homo sapiens, Marine actinobacteria clade
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2025-04-02 Classification: MEMBRANE PROTEIN Ligands: GDP, LFA, RET |
 |
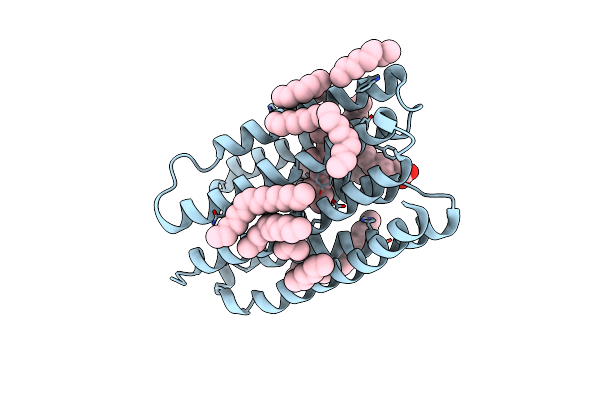
Crystal Structure Of Marine Actinobacteria Clade Rhodopsin (Mar) - Human Gtpase Arf1 (L8K,Q71L) Chimera; N State
Organism: Candidatus actinomarina minuta, Homo sapiens, Marine actinobacteria clade
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2025-04-02 Classification: MEMBRANE PROTEIN Ligands: GDP, LFA, OLA, RET |
 |
Crystal Structure Of Marine Actinobacteria Clade Rhodopsin (Mar) In The O* State
Organism: Candidatus actinomarina minuta, Marine actinobacteria clade
Method: X-RAY DIFFRACTION Resolution:1.41 Å Release Date: 2025-04-02 Classification: MEMBRANE PROTEIN Ligands: OLA, LFA, RET |
 |
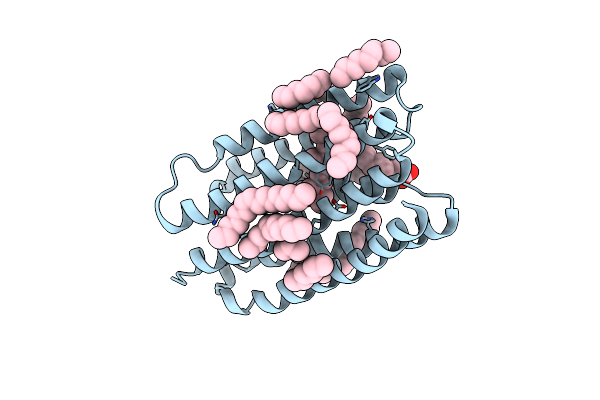
Crystal Structure Of Marine Actinobacteria Clade Rhodopsin (Mar) In The O* State, Ph 8.8
Organism: Candidatus actinomarina minuta, Marine actinobacteria clade
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2025-04-02 Classification: MEMBRANE PROTEIN Ligands: OLA, LFA, RET |
 |
Crystal Structure Of Marine Actinobacteria Clade Rhodopsin (Mar) In The O State Obtained By Cryotrapping
Organism: Candidatus actinomarina minuta, Marine actinobacteria clade
Method: X-RAY DIFFRACTION Resolution:1.51 Å Release Date: 2025-04-02 Classification: MEMBRANE PROTEIN Ligands: OLA, LFA, RET |
 |
Laser Excitation Effects On Br: Reprocessed Dark Dataset Recorded From Nango Et Al.
Organism: Halobacterium salinarum
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2025-01-22 Classification: PROTON TRANSPORT Ligands: LI1, OLC, RET |
 |
Laser Excitation Effects On Br: Reprocessed Extrapolated 16 Ns Light Dataset Recorded At 0.04 Gw/Cm2 From Nango Et Al.
Organism: Halobacterium salinarum
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2025-01-22 Classification: PROTON TRANSPORT Ligands: LI1, OLC, RET |
 |
Organism: Halobacterium salinarum
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2025-01-22 Classification: PROTON TRANSPORT Ligands: LI1, OLC, RET |
 |
Laser Excitation Effects On Br: Extrapolated 6 Ps Light Dataset Recorded At 342 Gw/Cm2 At Swissfel
Organism: Halobacterium salinarum
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2025-01-22 Classification: PROTON TRANSPORT Ligands: LI1, OLC, RET |
 |
Laser Excitation Effects On Br: Extrapolated 6 Ps Light Dataset Recorded At 2493 Gw/Cm2 At Swissfel
Organism: Halobacterium salinarum
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2025-01-22 Classification: PROTON TRANSPORT Ligands: LI1, OLC, RET |
 |
Organism: Halobacterium salinarum
Method: X-RAY DIFFRACTION Resolution:1.44 Å Release Date: 2025-01-22 Classification: PROTON TRANSPORT Ligands: LI1, OLC, RET |
 |
Laser Excitation Effects On Br: Reprocessed Extrapolated 10Ps Light Dataset Recorded At 525 Gw/Cm2 From Nogly Et Al.
Organism: Halobacterium salinarum
Method: X-RAY DIFFRACTION Resolution:1.66 Å Release Date: 2025-01-22 Classification: PROTON TRANSPORT Ligands: LI1, OLC, RET |

