Search Count: 515
All
Selected
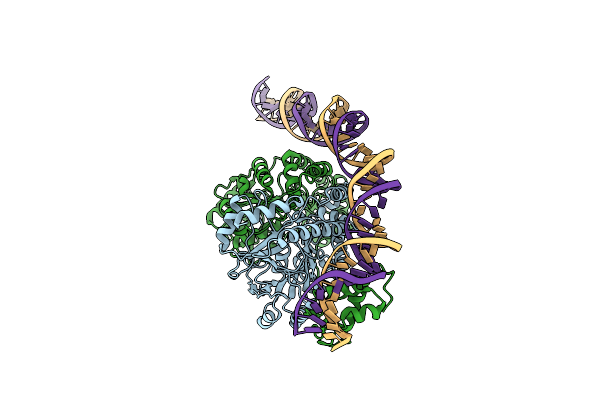 |
Cryo-Em Structure Of Holo-Pdxr From Bacillus Clausii Bound To Its Target Dna In The Half-Closed Conformation
Organism: Alkalihalobacillus clausii
Method: ELECTRON MICROSCOPY Release Date: 2023-07-05 Classification: DNA BINDING PROTEIN |
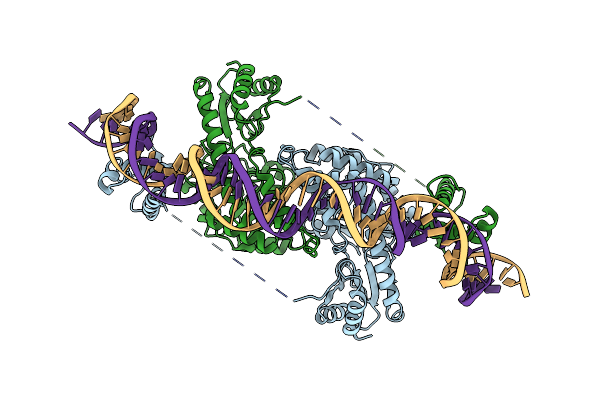 |
Cryo-Em Structure Of Holo-Pdxr From Bacillus Clausii Bound To Its Target Dna In The Closed Conformation, C2 Symmetry.
Organism: Alkalihalobacillus clausii
Method: ELECTRON MICROSCOPY Release Date: 2023-07-05 Classification: DNA BINDING PROTEIN |
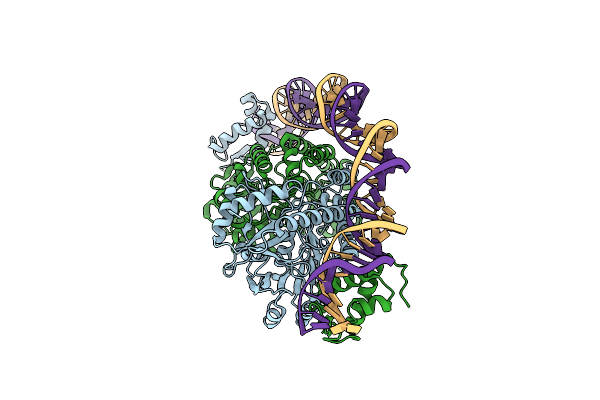 |
Cryo-Em Structure Of Holo-Pdxr From Bacillus Clausii Bound To Its Target Dna In The Closed Conformation, C1 Symmetry
Organism: Alkalihalobacillus clausii
Method: ELECTRON MICROSCOPY Release Date: 2023-07-05 Classification: DNA BINDING PROTEIN |
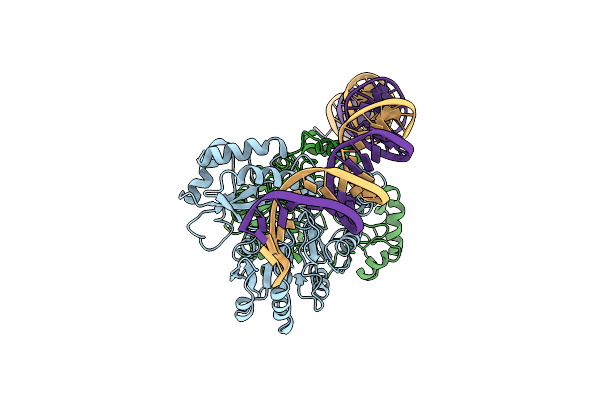 |
Cryo-Em Structure Of Holo-Pdxr From Bacillus Clausii Bound To Its Target Dna In The Open Conformation
Organism: Alkalihalobacillus clausii
Method: ELECTRON MICROSCOPY Release Date: 2023-07-05 Classification: DNA BINDING PROTEIN |
 |
Organism: Alkalihalobacillus clausii
Method: X-RAY DIFFRACTION Resolution:1.78 Å Release Date: 2023-03-29 Classification: LYASE Ligands: PEG, CA |
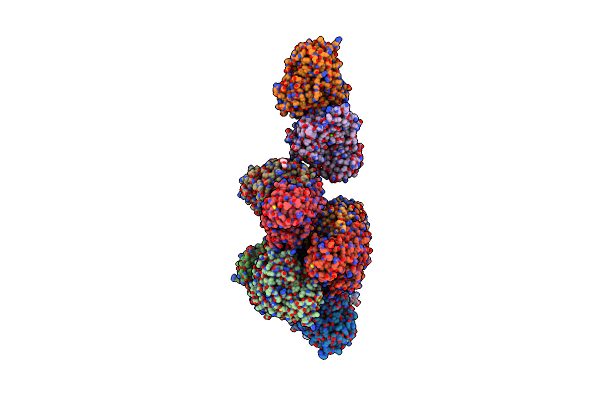 |
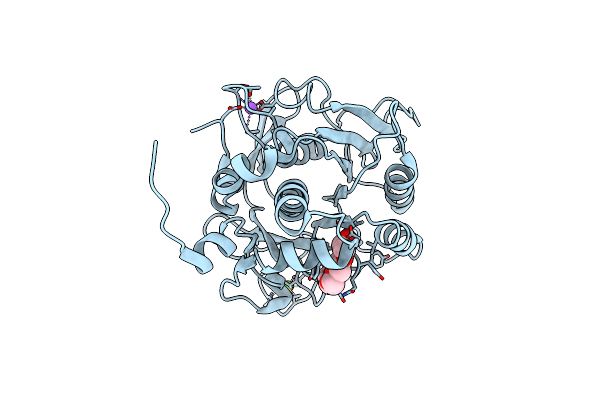
Organism: Alkalihalobacillus clausii
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2022-09-28 Classification: DNA BINDING PROTEIN Ligands: CA, CL, PGE, EDO, PEG, P6G |
 |
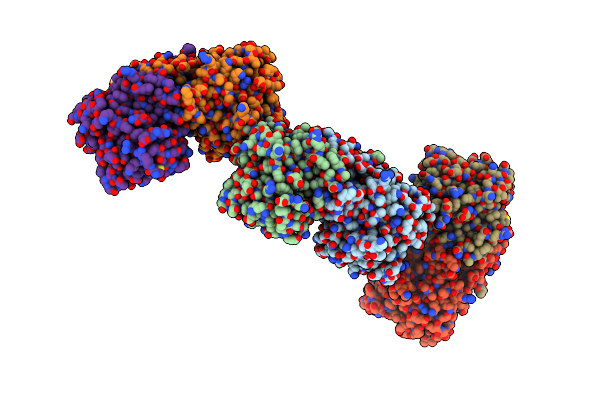
Organism: Bacillus clausii
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2011-03-16 Classification: HYDROLASE |
 |
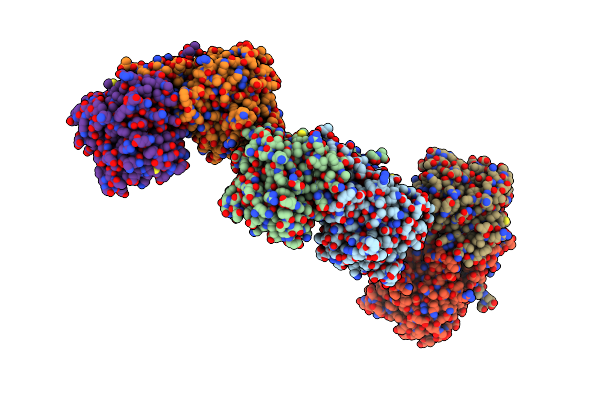
Organism: Bacillus clausii
Method: X-RAY DIFFRACTION Resolution:2.45 Å Release Date: 2010-09-08 Classification: HYDROLASE |
 |
Organism: Bacillus clausii
Method: X-RAY DIFFRACTION Resolution:2.68 Å Release Date: 2010-09-08 Classification: HYDROLASE |
 |
Organism: Bacillus clausii
Method: X-RAY DIFFRACTION Resolution:1.56 Å Release Date: 2010-08-11 Classification: HYDROLASE |
 |
Organism: Bacillus clausii ksm-k16
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2008-09-02 Classification: LYASE |
 |
Organism: Bacillus clausii
Method: X-RAY DIFFRACTION Resolution:1.82 Å Release Date: 2007-05-22 Classification: STRUCTURAL GENOMICS, UNKNOWN FUNCTION Ligands: EDO |
 |
Organism: Bacillus clausii
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2004-11-16 Classification: HYDROLASE Ligands: CA, SO4 |
 |
Crystal Structure Of A New Alkaline Serine Protease (M-Protease) From Bacillus Sp. Ksm-K16
Organism: Bacillus clausii
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 1994-06-22 Classification: SERINE PROTEINASE Ligands: CA |
 |
 |
NA
|
 |
NA
|
 |
NA
|
 |
NA
|
 |
NA
Organism: Bacillus clausii (strain KSM-K16)
Method: Alphafold Release Date: Classification: NA Ligands: NA |

