Search Count: 13
 |
Circularly Permuted Lumazine Synthase Twisted Tube With No Gap Between Between Double Strands
Organism: Aquifex aeolicus vf5
Method: ELECTRON MICROSCOPY Release Date: 2025-03-05 Classification: DE NOVO PROTEIN |
 |
Circularly Permuted Lumazine Synthase Twisted Tube With 18 Angstrom Gap Between Double Strands
Organism: Aquifex aeolicus vf5
Method: ELECTRON MICROSCOPY Release Date: 2025-03-05 Classification: DE NOVO PROTEIN |
 |
Circularly Permuted Lumazine Synthase Twisted Tube With 28 Angstrom Gap Between Double Strands
Organism: Aquifex aeolicus vf5
Method: ELECTRON MICROSCOPY Release Date: 2025-03-05 Classification: DE NOVO PROTEIN |
 |
Organism: Aquifex aeolicus vf5
Method: ELECTRON MICROSCOPY Release Date: 2025-03-05 Classification: DE NOVO PROTEIN |
 |
Organism: Aquifex aeolicus vf5
Method: ELECTRON MICROSCOPY Release Date: 2025-03-05 Classification: DE NOVO PROTEIN |
 |
Organism: Aquifex aeolicus vf5
Method: ELECTRON MICROSCOPY Release Date: 2025-03-05 Classification: DE NOVO PROTEIN |
 |
Organism: Aquifex aeolicus vf5
Method: ELECTRON MICROSCOPY Release Date: 2025-03-05 Classification: DE NOVO PROTEIN |
 |
Organism: Geobacillus stearothermophilus
Method: ELECTRON MICROSCOPY Release Date: 2024-12-25 Classification: VIRUS LIKE PARTICLE Ligands: CO |
 |
Structure Of The Co(Ii) Triggered Trap (S33Hk35H) Protein Cage (Dextro Form)
Organism: Geobacillus stearothermophilus
Method: ELECTRON MICROSCOPY Release Date: 2024-12-25 Classification: VIRUS LIKE PARTICLE Ligands: CO |
 |
Organism: Vicugna pacos
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2022-07-20 Classification: IMMUNE SYSTEM Ligands: SO4, EDO |
 |
Cryo-Em Structure Of The Sars-Cov-2 Spike Protein (2-Up Rbd) Bound To Neutralizing Nanobodies P86
Organism: Severe acute respiratory syndrome coronavirus 2, Vicugna pacos
Method: ELECTRON MICROSCOPY Release Date: 2022-07-20 Classification: VIRAL PROTEIN/IMMUNE SYSTEM Ligands: NAG |
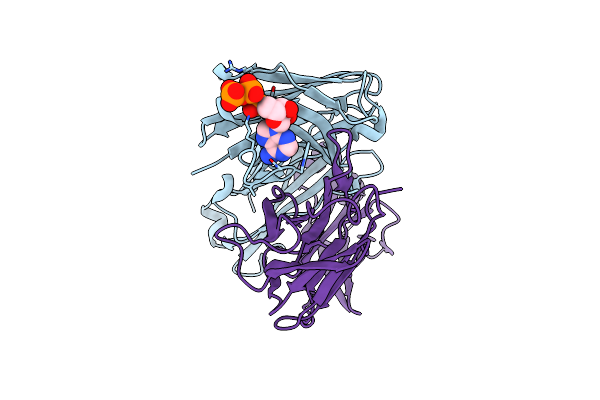 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.77 Å Release Date: 2021-01-13 Classification: IMMUNE SYSTEM Ligands: ATP |
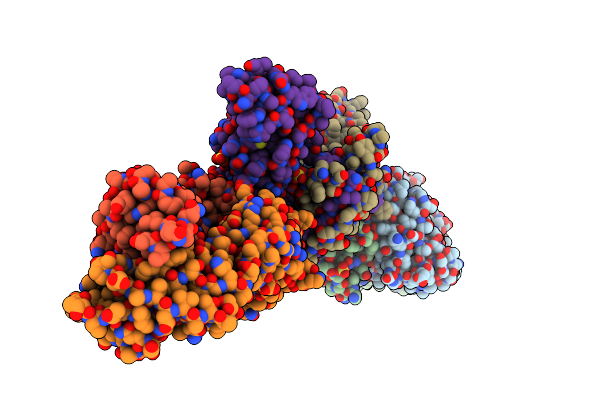 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.76 Å Release Date: 2021-01-13 Classification: IMMUNE SYSTEM Ligands: SO4, ATP |

