Search Count: 172
 |
Organism: Streptomyces lividans
Method: X-RAY DIFFRACTION Release Date: 2025-09-03 Classification: OXIDOREDUCTASE Ligands: HEM, NO |
 |
Organism: Streptomyces lividans
Method: X-RAY DIFFRACTION Release Date: 2025-09-03 Classification: OXIDOREDUCTASE Ligands: HEM, NO |
 |
Sfx Structure Of Cytochrome C Prime Beta From Methylococcus Capsulatus (Bath)
Organism: Methylococcus capsulatus str. bath
Method: X-RAY DIFFRACTION Release Date: 2025-09-03 Classification: METAL BINDING PROTEIN Ligands: HEC, GOL, ZN, SO4 |
 |
Cytochrome C Prime Beta From Methylococcus Capsulatus (Bath) In Complex With Nitric Oxide From Proli Nonoate
Organism: Methylococcus capsulatus str. bath
Method: X-RAY DIFFRACTION Release Date: 2025-09-03 Classification: METAL BINDING PROTEIN Ligands: HEC, GOL, ZN, NO |
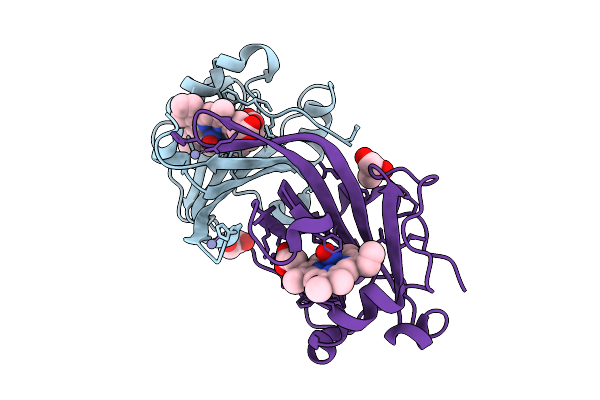 |
Organism: Methylococcus capsulatus str. bath
Method: X-RAY DIFFRACTION Release Date: 2025-09-03 Classification: METAL BINDING PROTEIN Ligands: HEC, GOL, NO, ZN |
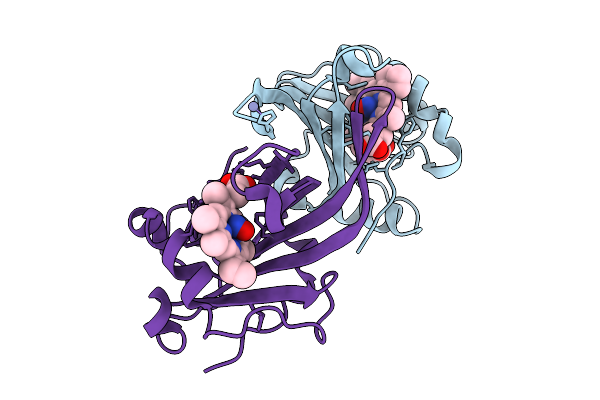 |
Organism: Methylococcus capsulatus str. bath
Method: X-RAY DIFFRACTION Release Date: 2025-09-03 Classification: METAL BINDING PROTEIN Ligands: HEC, ZN, NO |
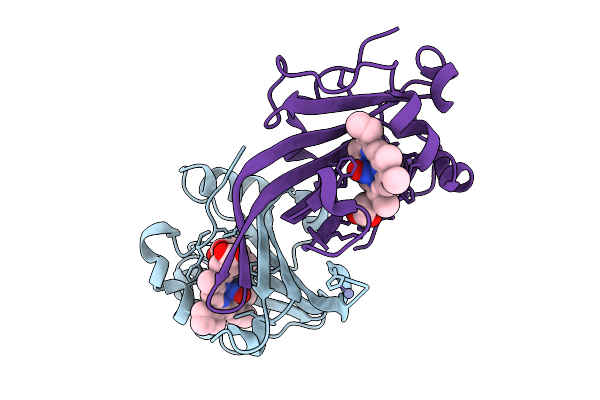 |
Organism: Methylococcus capsulatus str. bath
Method: X-RAY DIFFRACTION Release Date: 2025-09-03 Classification: METAL BINDING PROTEIN Ligands: HEC, ZN, NO |
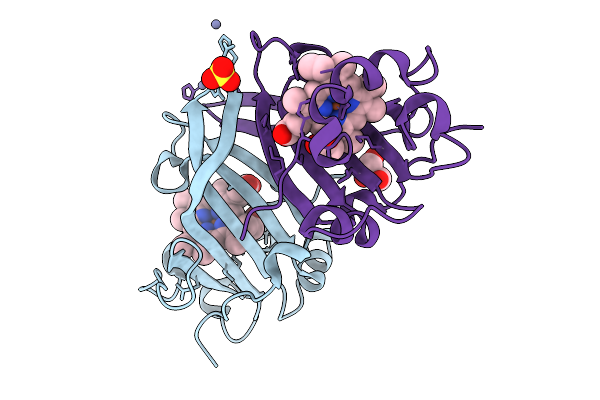 |
Ssx Structure Of Cytochrome C Prime Beta From Methylococcus Capsulatus (Bath)
Organism: Methylococcus capsulatus str. bath
Method: X-RAY DIFFRACTION Release Date: 2025-09-03 Classification: METAL BINDING PROTEIN Ligands: HEC, ZN, GOL, SO4 |
 |
Dtpb In Complex With Photocaged Nitric Oxide, 100 Microsecond, 30 Microjoule, Sfx
Organism: Streptomyces lividans 1326
Method: X-RAY DIFFRACTION Release Date: 2025-09-03 Classification: OXIDOREDUCTASE Ligands: HEM, NO |
 |
Dtpb In Complex With Photocaged Nitric Oxide, 100 Microsecond, 100 Microjoule, Sfx
Organism: Streptomyces lividans
Method: X-RAY DIFFRACTION Release Date: 2025-09-03 Classification: OXIDOREDUCTASE Ligands: HEM, NO2 |
 |
Organism: Methylococcus capsulatus str. bath
Method: X-RAY DIFFRACTION Release Date: 2025-09-03 Classification: METAL BINDING PROTEIN Ligands: HEC, ZN, GOL |
 |
Organism: Streptomyces lividans 1326
Method: X-RAY DIFFRACTION Release Date: 2025-09-03 Classification: METAL BINDING PROTEIN Ligands: HEM, MG |
 |
Organism: Streptomyces lividans 1326
Method: X-RAY DIFFRACTION Release Date: 2025-09-03 Classification: METAL BINDING PROTEIN Ligands: HEM, NO |
 |
Organism: Streptomyces lividans 1326
Method: X-RAY DIFFRACTION Release Date: 2025-09-03 Classification: METAL BINDING PROTEIN Ligands: HEM, NO |
 |
Organism: Streptomyces lividans 1326
Method: X-RAY DIFFRACTION Release Date: 2025-09-03 Classification: METAL BINDING PROTEIN Ligands: HEM, NO |
 |
Organism: Streptomyces lividans 1326
Method: X-RAY DIFFRACTION Release Date: 2025-09-03 Classification: METAL BINDING PROTEIN Ligands: HEM, NO |
 |
Organism: Streptomyces lividans 1326
Method: X-RAY DIFFRACTION Release Date: 2025-09-03 Classification: METAL BINDING PROTEIN Ligands: HEM, NO |
 |
Organism: Methylococcus capsulatus str. bath
Method: X-RAY DIFFRACTION Release Date: 2025-09-03 Classification: METAL BINDING PROTEIN Ligands: HEC, ZN, NO |
 |
Organism: Methylococcus capsulatus str. bath
Method: X-RAY DIFFRACTION Release Date: 2025-09-03 Classification: METAL BINDING PROTEIN Ligands: ZN, HEC, NO, GOL |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2024-04-17 Classification: STRUCTURAL PROTEIN |

