Search Count: 14
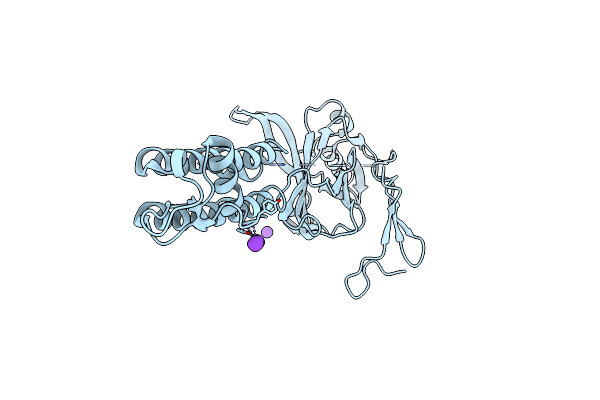 |
Crystal Structure Of The Eukaryotic Strong Inward-Rectifier K+ Channel Kir2.2 At 3.1 Angstrom Resolution
Organism: Gallus gallus
Method: X-RAY DIFFRACTION Resolution:3.11 Å Release Date: 2010-01-12 Classification: METAL TRANSPORT Ligands: K |
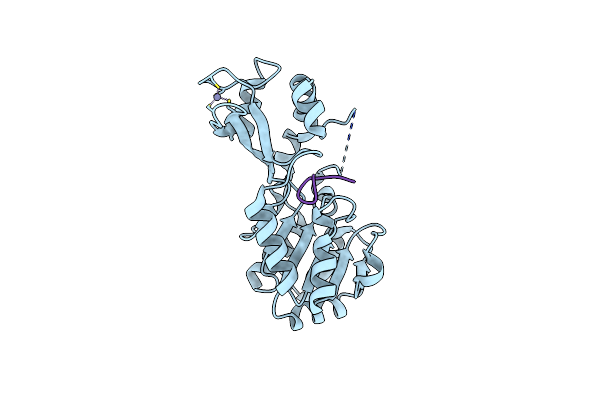 |
Organism: Thermotoga maritima
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2006-12-05 Classification: HYDROLASE Ligands: ZN |
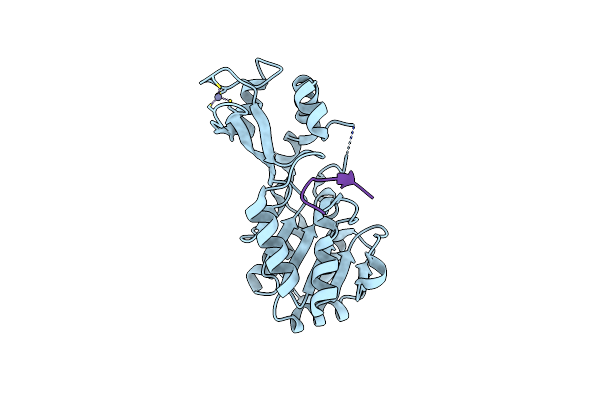 |
Organism: Thermotoga maritima
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2006-12-05 Classification: HYDROLASE Ligands: ZN |
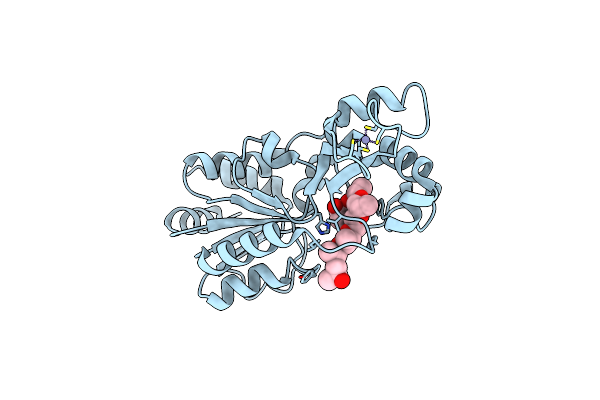 |
Organism: Thermotoga maritima
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2006-12-05 Classification: HYDROLASE Ligands: ZN, ZPG |
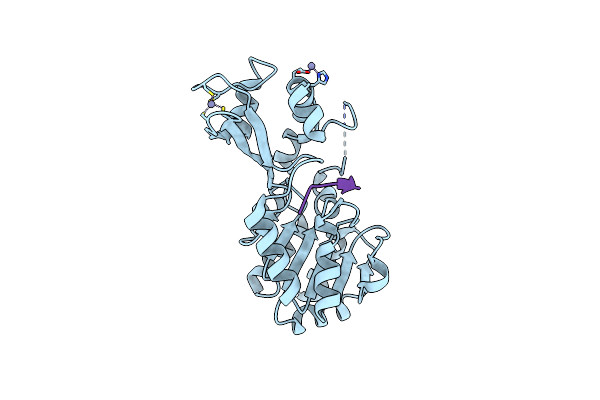 |
Organism: Thermotoga maritima
Method: X-RAY DIFFRACTION Resolution:1.63 Å Release Date: 2006-11-28 Classification: HYDROLASE Ligands: ZN |
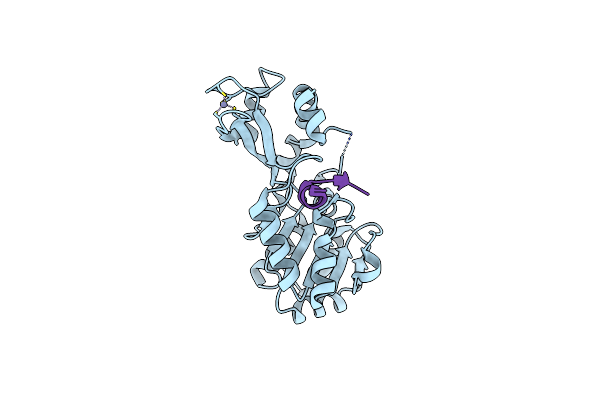 |
Organism: Thermotoga maritima
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2006-09-19 Classification: HYDROLASE Ligands: ZN |
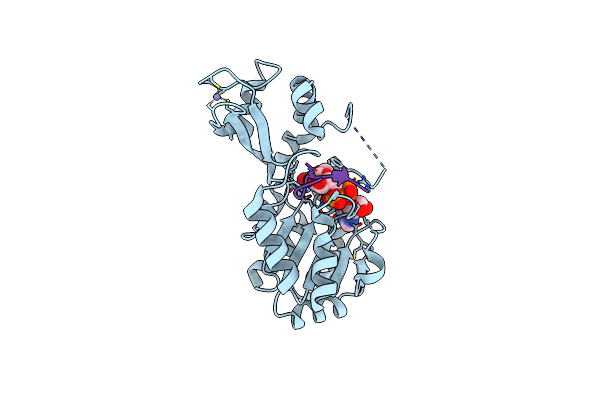 |
Organism: Thermotoga maritima
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2006-09-05 Classification: HYDROLASE Ligands: ZN, NAD |
 |
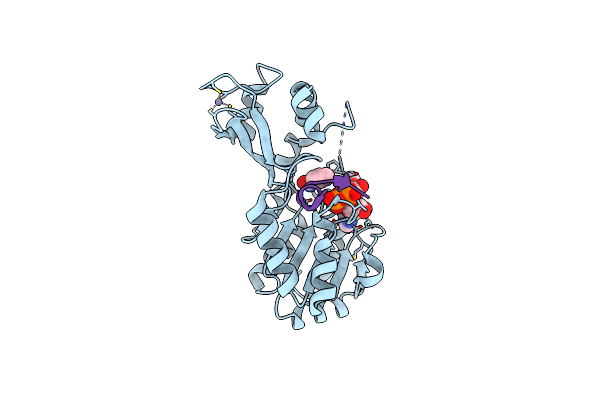
Organism: Thermotoga maritima
Method: X-RAY DIFFRACTION Resolution:1.99 Å Release Date: 2006-09-05 Classification: HYDROLASE Ligands: ZN, NAD |
 |
Organism: Thermotoga maritima
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2006-09-05 Classification: HYDROLASE Ligands: ZN, NCA, OAD |
 |
Organism: Thermotoga maritima
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2006-09-05 Classification: HYDROLASE Ligands: ZN, APR, 3OD |
 |
Organism: Thermotoga maritima
Method: X-RAY DIFFRACTION Resolution:1.40 Å Release Date: 2005-04-26 Classification: HYDROLASE Ligands: ZN, NCA |
 |
Organism: Archaeoglobus fulgidus
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2005-03-29 Classification: HYDROLASE Ligands: ZN, SO4, NAD, NCA, EDO, 2PE, PG4, PGE, APR |
 |
Organism: Archaeoglobus fulgidus
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2004-03-23 Classification: TRANSCRIPTION Ligands: ZN, SO4, NAD, 1PE, EDO, 2PE, P6G, PG4, APR |
 |
Organism: Archaeoglobus fulgidus
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2002-10-16 Classification: PROTEIN BINDING, TRANSCRIPTION Ligands: ZN, MES |

