Search Count: 35
 |
Room-Temperature Serial Synchrotron Crystallography Structure Of Spinacia Oleracea Rubisco
Organism: Spinacia oleracea
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2024-05-29 Classification: LYASE Ligands: MG |
 |
Ribonucleotide Reductase Class Ie R2 From Mesoplasma Florum, Catalytically Active Radical State Solved By Xfel
Organism: Mesoplasma florum l1
Method: X-RAY DIFFRACTION Release Date: 2023-11-01 Classification: OXIDOREDUCTASE |
 |
Ribonucleotide Reductase Class Ie R2 From Mesoplasma Florum, Radical-Lost Ground State
Organism: Mesoplasma florum l1
Method: X-RAY DIFFRACTION Release Date: 2023-11-01 Classification: OXIDOREDUCTASE Ligands: CA, GOL |
 |
Methemoglobin Structure From Serial Synchrotron Crystallography With Fixed Target
Organism: Equus caballus
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2023-10-18 Classification: OXYGEN TRANSPORT Ligands: HEM |
 |
Dexyhemoglobin Structure From Serial Synchrotron Crystallography With Fixed Target
Organism: Equus caballus
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2023-10-18 Classification: OXYGEN TRANSPORT Ligands: HEM |
 |
Determination Of The Structure Of Active Tyrosinase From Bacterium Verrucomicrobium Spinosum
Organism: Verrucomicrobium spinosum
Method: X-RAY DIFFRACTION Resolution:1.43 Å Release Date: 2023-09-20 Classification: METAL BINDING PROTEIN Ligands: GOL, SO4, CU |
 |
Determination Of The Structure Of Active Tyrosinase From Bacterium Verrucomicrobium Spinosum
Organism: Verrucomicrobium spinosum
Method: X-RAY DIFFRACTION Resolution:1.64 Å Release Date: 2023-09-20 Classification: METAL BINDING PROTEIN Ligands: CU, SO4 |
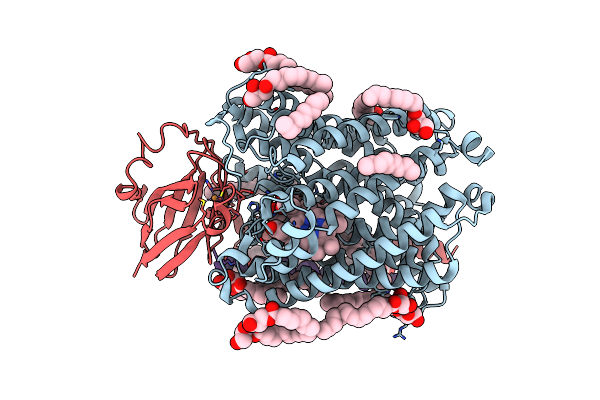 |
Serial Synchrotron Crystallography Structure Of Ba3-Type Cytochrome C Oxidase From Thermus Thermophilus Using A Goniometer Compatible Flow-Cell
Organism: Thermus thermophilus, Thermus thermophilus hb8
Method: X-RAY DIFFRACTION Resolution:2.12 Å Release Date: 2023-03-01 Classification: ELECTRON TRANSPORT Ligands: CU, HEM, HAS, OLC, CUA |
 |
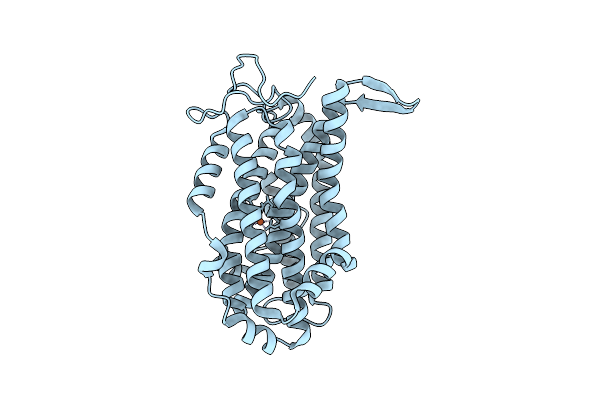
Xfel Structure Of Class Ib Ribonucleotide Reductase Dimanganese(Ii) Nrdf In Complex With Oxidized Nrdi From Bacillus Cereus
Organism: Bacillus cereus atcc 14579
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2022-09-21 Classification: OXIDOREDUCTASE Ligands: MN, UNX, FMN |
 |
Xfel Structure Of Class Ib Ribonucleotide Reductase Dimanganese(Ii) Nrdf In Complex With Hydroquinone Nrdi From Bacillus Cereus
Organism: Bacillus cereus atcc 14579
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2022-09-21 Classification: OXIDOREDUCTASE Ligands: MN, FNR, UNX |
 |
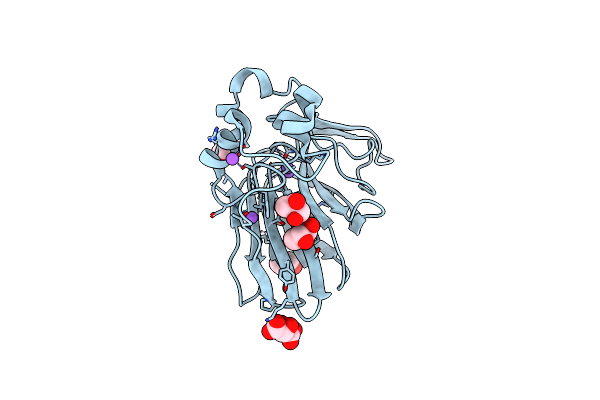
Structure Of Ribonucleotide Reductase R2 From Escherichia Coli Collected By Femtosecond Serial Crystallography On A Coc Membrane
Organism: Escherichia coli (strain k12)
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2022-01-12 Classification: OXIDOREDUCTASE Ligands: FE |
 |
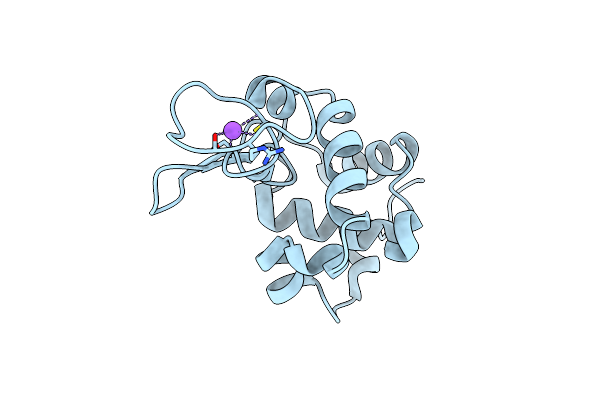
Structure Of Thaumatin Collected By Femtosecond Serial Crystallography On A Coc Membrane
Organism: Thaumatococcus daniellii
Method: X-RAY DIFFRACTION Resolution:1.46 Å Release Date: 2021-11-24 Classification: PLANT PROTEIN Ligands: TLA, PGR, NA |
 |
Structure Of Hen Egg White Lysozyme Collected By Rotation Serial Crystallography On A Coc Membrane At A Synchrotron Source
Organism: Gallus gallus
Method: X-RAY DIFFRACTION Resolution:1.51 Å Release Date: 2021-09-01 Classification: HYDROLASE Ligands: CL, NA |
 |
Structure Of Thaumatin Collected By Rotation Serial Crystallography On A Coc Membrane At A Synchrotron Source
Organism: Thaumatococcus daniellii
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2021-09-01 Classification: PLANT PROTEIN Ligands: TLA, PGO, NA |
 |
Structure Of Insulin Collected By Rotation Serial Crystallography On A Coc Membrane At A Synchrotron Source
Organism: Sus scrofa
Method: X-RAY DIFFRACTION Resolution:1.46 Å Release Date: 2021-09-01 Classification: HORMONE Ligands: PGR, NA |
 |
Structure Of Tubulin Darpin Complex 1 Collected By Rotation Serial Crystallography On A Coc Membrane At A Synchrotron Source
Organism: Bos taurus
Method: X-RAY DIFFRACTION Resolution:2.26 Å Release Date: 2021-09-01 Classification: CELL CYCLE Ligands: GTP, MG, GDP, PG0 |
 |
Structure Of Sponge-Phase Grown Peptst2 Collected By Rotation Serial Crystallography On A Coc Membrane At A Synchrotron Source
Organism: Streptococcus thermophilus (strain atcc baa-250 / lmg 18311)
Method: X-RAY DIFFRACTION Resolution:3.00 Å Release Date: 2021-09-01 Classification: TRANSPORT PROTEIN Ligands: PO4, 78M, PG0, PGE |
 |
Structure Of Ribonucleotide Reductase R2 From Escherichia Coli Collected By Still Serial Crystallography On A Coc Membrane At A Synchrotron Source
Organism: Escherichia coli (strain k12)
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2021-09-01 Classification: OXIDOREDUCTASE Ligands: FE |
 |
Structure Of Ribonucleotide Reductase R2 From Escherichia Coli Collected By Rotation Serial Crystallography On A Coc Membrane At A Synchrotron Source
Organism: Escherichia coli (strain k12)
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2021-09-01 Classification: OXIDOREDUCTASE Ligands: FE |
 |
Crystal Structure Of Ribonucleotide Reductase R2 Subunit Solved By Serial Synchrotron Crystallography
Organism: Saccharopolyspora erythraea (strain atcc 11635 / dsm 40517 / jcm 4748 / nbrc 13426 / ncimb 8594 / nrrl 2338)
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2020-10-07 Classification: OXIDOREDUCTASE Ligands: MN3, FE |

