Search Count: 16
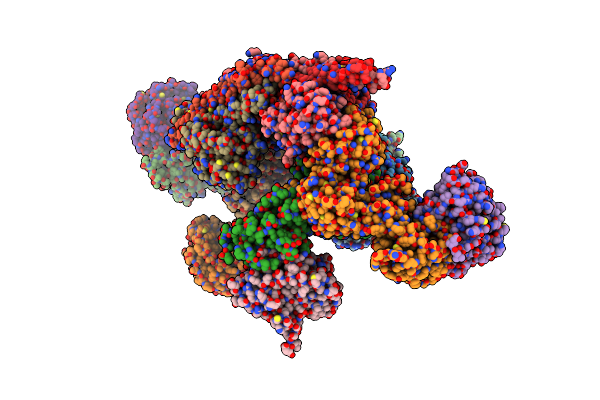 |
Cryo-Em Structure Of The Human Brisc Dimer Complex Bound To Compound Fx-171-C
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-02-12 Classification: SIGNALING PROTEIN Ligands: ZN, G1V |
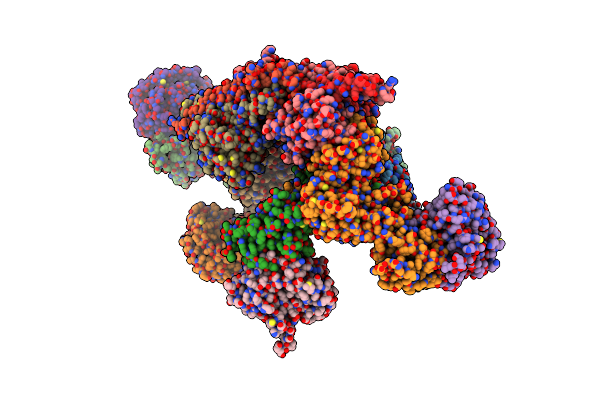 |
Cryo-Em Structure Of The Human Brisc Dimer Complex Bound To Compound Jms-175-2
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-02-05 Classification: SIGNALING PROTEIN Ligands: ZN, X8C |
 |
Organism: Homo sapiens, Bos taurus
Method: ELECTRON MICROSCOPY Release Date: 2024-05-29 Classification: CELL CYCLE |
 |
Organism: Homo sapiens, Lama glama
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2023-09-27 Classification: STRUCTURAL PROTEIN Ligands: EDO, PEG |
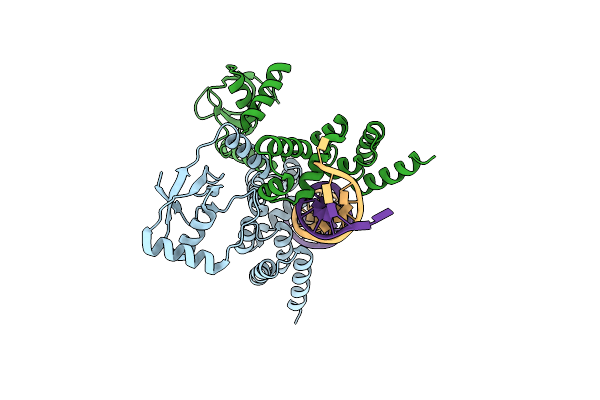 |
Crystal Structure Of C-Terminally Truncated Bacillus Subtilis Nucleoid Occlusion Protein (Noc) Complexed To The Noc-Binding Site (Nbs)
Organism: Bacillus subtilis (strain 168), Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.90 Å Release Date: 2022-03-09 Classification: DNA BINDING PROTEIN |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.43 Å Release Date: 2021-01-27 Classification: CELL CYCLE |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2019-06-05 Classification: SIGNALING PROTEIN Ligands: ZN |
 |
Structure Of A Modified Protein Containing A Genetically Encoded Phosphoserine
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.66 Å Release Date: 2018-10-24 Classification: CHAPERONE Ligands: ADP, PO4, MG, K |
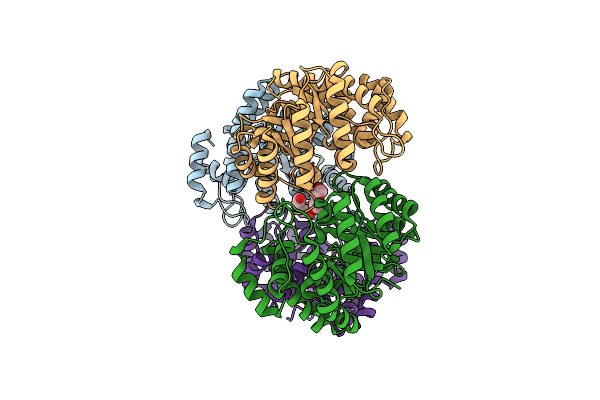 |
X-Ray Crystal Structure Of N-Acetylneuraminic Acid Lyase In Complex With Pyruvate, With The Phenylalanine At Position 190 Replaced With The Non-Canonical Amino Acid Dihydroxypropylcysteine.
Organism: Staphylococcus aureus (strain nctc 8325)
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2017-03-22 Classification: LYASE Ligands: PEG |
 |
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Resolution:1.99 Å Release Date: 2014-06-18 Classification: HYDROLASE/HYDROLASE INHIBITOR Ligands: EDO, A08 |
 |
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2014-06-18 Classification: HYDROLASE/HYDROLASE INHIBITOR Ligands: EDO, 1OQ |
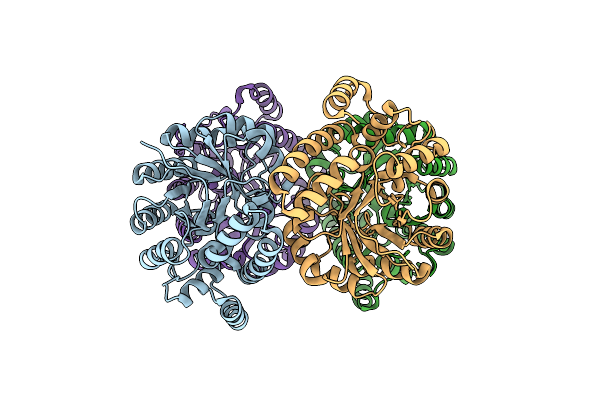 |
Structure Of Wild Type Stapylococcus Aureus N-Acetylneuraminic Acid Lyase In Complex With Pyruvate
Organism: Staphylococcus aureus subsp. aureus nctc 8325
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2013-01-23 Classification: LYASE |
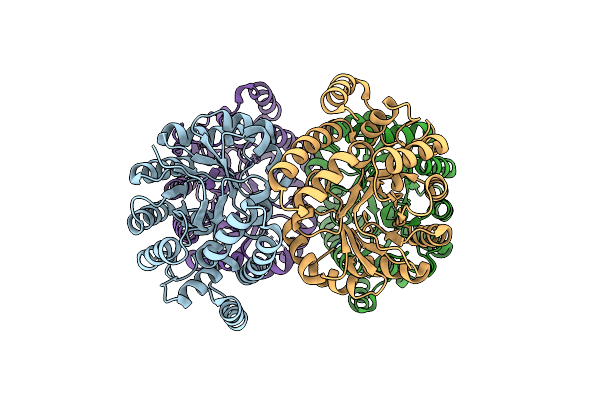 |
Crystal Structure Of N-Acetylneuraminic Acid Lyase From Staphylococcus Aureus With The Chemical Modification Thia-Lysine At Position 165
Organism: Staphylococcus aureus subsp. aureus nctc 8325
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2013-01-23 Classification: LYASE Ligands: CL |
 |
Crystal Structure Of Wild Type N-Acetylneuraminic Acid Lyase From Staphylococcus Aureus
Organism: Staphylococcus aureus subsp. aureus nctc 8325
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2013-01-23 Classification: LYASE Ligands: CL |
 |
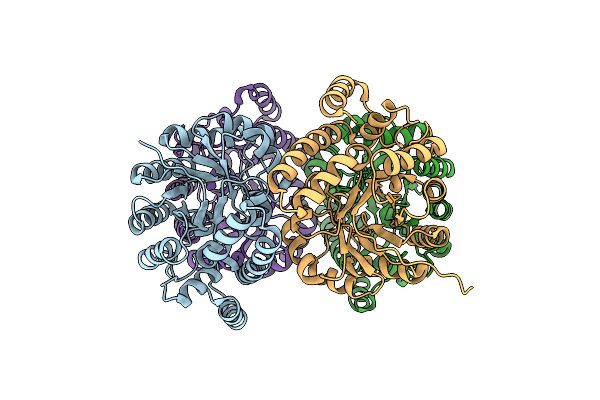
Crystal Structure Of N-Acetylneuraminic Acid Lyase Mutant K165C From Staphylococcus Aureus
Organism: Staphylococcus aureus subsp. aureus nctc 8325
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2013-01-23 Classification: LYASE |
 |
Crystal Structure Of N-Acetylneuraminic Acid Lyase From Staphylococcus Aureus With The Chemical Modification Thia-Lysine At Position 165 In Complex With Pyruvate
Organism: Staphylococcus aureus subsp. aureus nctc 8325
Method: X-RAY DIFFRACTION Resolution:2.35 Å Release Date: 2013-01-23 Classification: LYASE |

