Search Count: 13
 |
Organism: Deinococcus radiodurans r1
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2014-10-01 Classification: FLUORESCENT PROTEIN Ligands: LBV, LBW |
 |
Organism: Unidentified
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2014-05-21 Classification: OXIDOREDUCTASE Ligands: FAD, MG, CL |
 |
Organism: Deinococcus radiodurans
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2013-12-25 Classification: SIGNALING PROTEIN Ligands: LBV, GOL, PO4, PEG |
 |
Organism: Lycopersicon hirsutum f. glabratum
Method: X-RAY DIFFRACTION Resolution:2.24 Å Release Date: 2012-05-02 Classification: HYDROLASE Ligands: DKA |
 |
Crystal Structure Of Tomato Methylketone Synthase I Complexed With Methyl-3-Hydroxydodecanoate
Organism: Lycopersicon hirsutum f. glabratum
Method: X-RAY DIFFRACTION Resolution:1.93 Å Release Date: 2012-05-02 Classification: HYDROLASE Ligands: DKA, DK3 |
 |
Crystal Structure Of Tomato Methylketone Synthase I Complexed With 3-Hydroxyoctanoate
Organism: Lycopersicon hirsutum f. glabratum
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2012-05-02 Classification: HYDROLASE Ligands: 3HO, BR |
 |
Crystal Structure Of Tomato Methylketone Synthase I Complexed With 2-Tridecanone
Organism: Lycopersicon hirsutum f. glabratum
Method: X-RAY DIFFRACTION Resolution:2.31 Å Release Date: 2012-05-02 Classification: HYDROLASE Ligands: 2TD |
 |
Crystal Structure Of Tomato Methylketone Synthase I H243A Variant Complexed With Beta-Ketoheptanoate
Organism: Lycopersicon hirsutum f. glabratum
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2012-05-02 Classification: HYDROLASE Ligands: BKA |
 |
Organism: Lycopersicon hirsutum f. glabratum
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2012-05-02 Classification: HYDROLASE Ligands: DKA |
 |
Crystal Structure Of A Monomeric Infrared Fluorescent Deinococcus Radiodurans Bacteriophytochrome Chromophore Binding Domain
Organism: Deinococcus radiodurans
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2012-01-11 Classification: FLUORESCENT PROTEIN Ligands: LBV, LBW, GOL, PO4 |
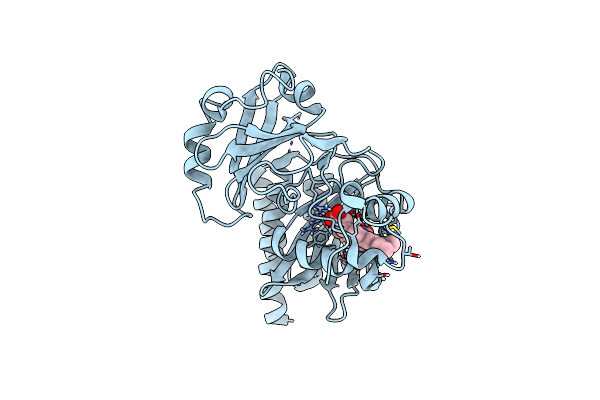 |
Crystal Structure Of The Alternate His 207 Conformation Of The Infrared Fluorescent D207H Variant Of Deinococcus Bacteriophytochrome Chromophore Binding Domain At 2.45 Angstrom Resolution
Organism: Deinococcus radiodurans
Method: X-RAY DIFFRACTION Resolution:2.45 Å Release Date: 2011-12-21 Classification: FLUORESCENT PROTEIN Ligands: LBV |
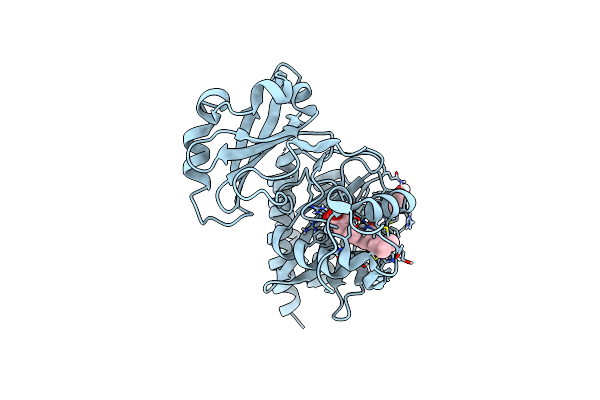 |
Crystal Structure Of The Infrared Fluorescent D207H Variant Of Deinococcus Bacteriophytochrome Chromophore Binding Domain At 1.24 Angstrom Resolution
Organism: Deinococcus radiodurans
Method: X-RAY DIFFRACTION Resolution:1.24 Å Release Date: 2011-12-21 Classification: FLUORESCENT PROTEIN Ligands: LBV, GOL |
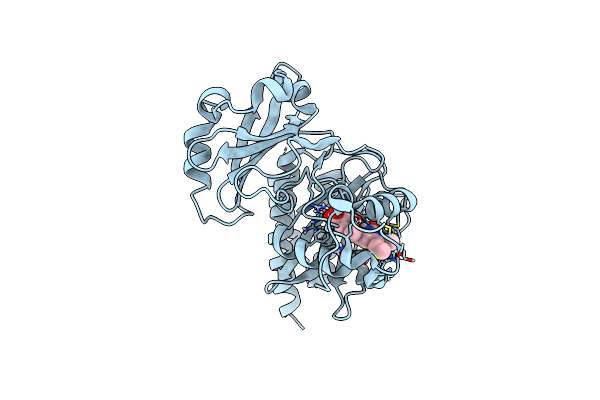 |
Crystal Structure Of The Infrared Fluorescent D207H Variant Of Deinococcus Bacteriophytochrome Chromophore Binding Domain At 1.72 Angstrom Resolution
Organism: Deinococcus radiodurans
Method: X-RAY DIFFRACTION Resolution:1.72 Å Release Date: 2011-12-21 Classification: FLUORESCENT PROTEIN Ligands: LBV |

