Search Count: 159
 |
Organism: Pseudomonas aeruginosa
Method: ELECTRON MICROSCOPY Release Date: 2025-11-19 Classification: LIPID TRANSPORT Ligands: 3PE |
 |
Organism: Pseudomonas aeruginosa
Method: ELECTRON MICROSCOPY Release Date: 2025-11-19 Classification: LIPID TRANSPORT Ligands: 3PE |
 |
Organism: Pseudomonas aeruginosa
Method: ELECTRON MICROSCOPY Release Date: 2025-11-19 Classification: LIPID TRANSPORT Ligands: 3PE |
 |
Organism: Pseudomonas aeruginosa
Method: ELECTRON MICROSCOPY Release Date: 2025-11-19 Classification: LIPID TRANSPORT Ligands: 3PE |
 |
Organism: Pseudomonas aeruginosa
Method: ELECTRON MICROSCOPY Release Date: 2025-11-19 Classification: LIPID TRANSPORT |
 |
Organism: Gallus gallus
Method: X-RAY DIFFRACTION Release Date: 2025-11-05 Classification: HYDROLASE Ligands: A1ISW, PEG, EDO, IMD, NA, CL |
 |
Organism: Aspergillus niger
Method: X-RAY DIFFRACTION Release Date: 2025-11-05 Classification: OXIDOREDUCTASE Ligands: EDO, MAN, NAG, FAD, CL, NA, BA, BR |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2024-05-08 Classification: HYDROLASE Ligands: ADP |
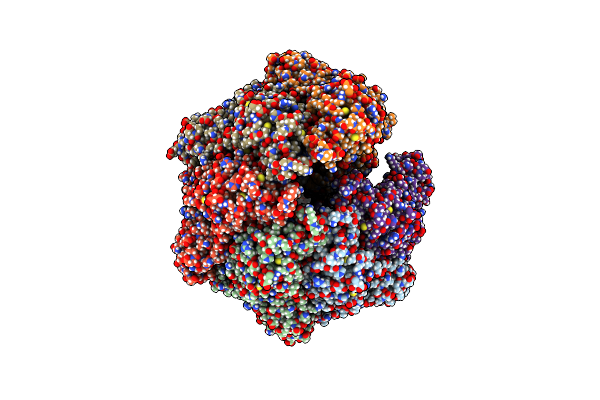 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2024-05-08 Classification: HYDROLASE |
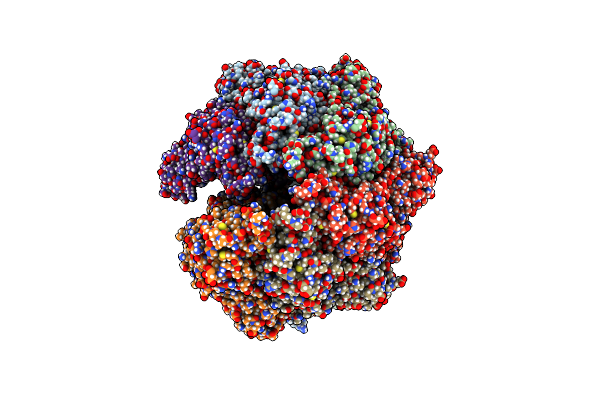 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2024-04-10 Classification: HYDROLASE Ligands: ADP |
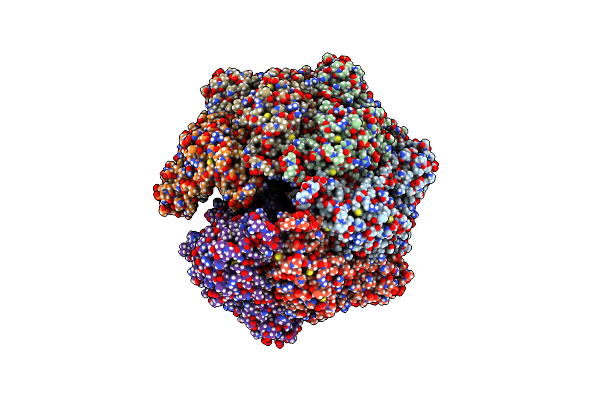 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2024-04-10 Classification: HYDROLASE Ligands: ADP |
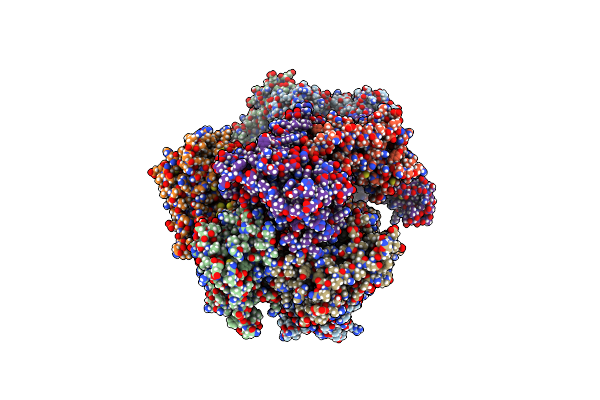 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2024-04-03 Classification: HYDROLASE Ligands: ADP |
 |
Organism: Arabidopsis thaliana
Method: X-RAY DIFFRACTION Resolution:2.52 Å Release Date: 2023-11-08 Classification: HYDROLASE Ligands: APR |
 |
Organism: Blumeria hordei
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2023-08-16 Classification: UNKNOWN FUNCTION Ligands: HOH |
 |
Crystal Structure Of Powdery Mildews Blumeria Graminis F. Sp. Tritici Avrpm2(1)
Organism: Blumeria graminis f. sp. tritici
Method: X-RAY DIFFRACTION Resolution:1.51 Å Release Date: 2023-08-16 Classification: UNKNOWN FUNCTION |
 |
Crystal Structure Of Powdery Mildews Blumeria Graminis F. Sp. Hordei Avra22
Organism: Blumeria hordei
Method: X-RAY DIFFRACTION Resolution:1.64 Å Release Date: 2023-08-16 Classification: UNKNOWN FUNCTION |
 |
Crystal Structure Of Powdery Mildews Blumeria Graminis F. Sp. Hordei Avra10
Organism: Blumeria hordei
Method: X-RAY DIFFRACTION Resolution:1.37 Å Release Date: 2023-08-16 Classification: UNKNOWN FUNCTION |
 |
Organism: Blumeria hordei dh14
Method: X-RAY DIFFRACTION Resolution:1.56 Å Release Date: 2023-08-16 Classification: UNKNOWN FUNCTION |
 |
Crystal Structure Of Powdery Mildews Blumeria Graminis F. Sp. Tritici Avrpm2 (2)
Organism: Blumeria graminis f. sp. tritici
Method: X-RAY DIFFRACTION Resolution:1.06 Å Release Date: 2023-08-16 Classification: UNKNOWN FUNCTION |
 |
Crystal Structure Of Ap2 Associated Kinase 1 Complexed With 5-[(4-Aminopiperidin-1-Yl)Methyl]-N-{3-[5-(Propan-2-Yl)-1,3,4-Thiadiazol-2-Yl]Phenyl}Pyrrolo[2,1-F][1,2,4]Triazin-4-Amine
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2023-07-12 Classification: TRANSFERASE Ligands: YFV, SO4 |

