Search Count: 22
 |
High-Resolution Crystal Structure Of Fluoropropylated Cystine Knot, Binding To Alpha-5 Beta-6 Integrin
Organism: Momordica cochinchinensis
Method: X-RAY DIFFRACTION Resolution:1.00 Å Release Date: 2019-08-14 Classification: PROTEIN BINDING |
 |
Organism: Mus musculus
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2015-09-02 Classification: IMMUNE SYSTEM |
 |
Organism: Mus musculus
Method: X-RAY DIFFRACTION Resolution:2.85 Å Release Date: 2015-09-02 Classification: IMMUNE SYSTEM Ligands: QI9 |
 |
Organism: Mus musculus
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2015-09-02 Classification: IMMUNE SYSTEM Ligands: SO4 |
 |
Organism: Mus musculus
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2015-09-02 Classification: IMMUNE SYSTEM Ligands: QI9 |
 |
Organism: Sorangium cellulosum
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2015-03-11 Classification: TRANSFERASE |
 |
Organism: Sorangium cellulosum
Method: X-RAY DIFFRACTION Resolution:2.47 Å Release Date: 2015-03-11 Classification: TRANSFERASE Ligands: IOD, GOL |
 |
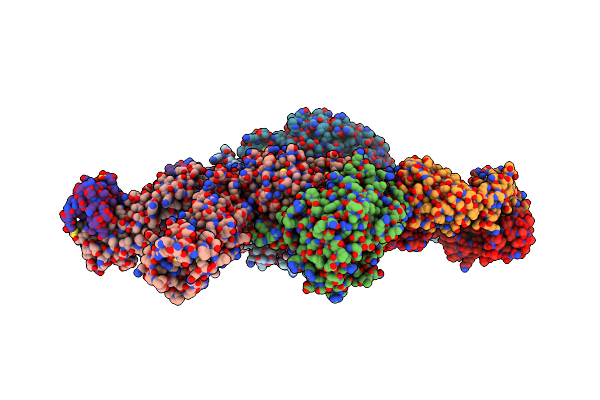
Crystal Structure Of I-Crei Complexed With Its Target Methylated At Position Plus 2 (In The B Strand) In The Presence Of Calcium
Organism: Chlamydomonas reinhardtii, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2012-07-04 Classification: HYDROLASE Ligands: CA, GOL |
 |
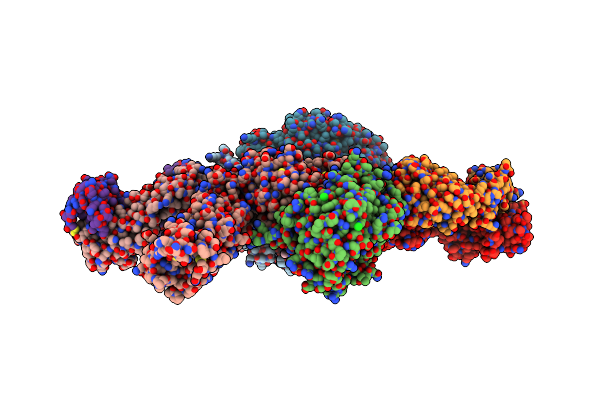
Crystal Structure Of I-Crei Complexed With Its Target Methylated At Position Plus 2 (In The B Strand) In The Presence Of Magnesium
Organism: Chlamydomonas reinhardtii, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2012-07-04 Classification: HYDROLASE Ligands: GOL, MG |
 |
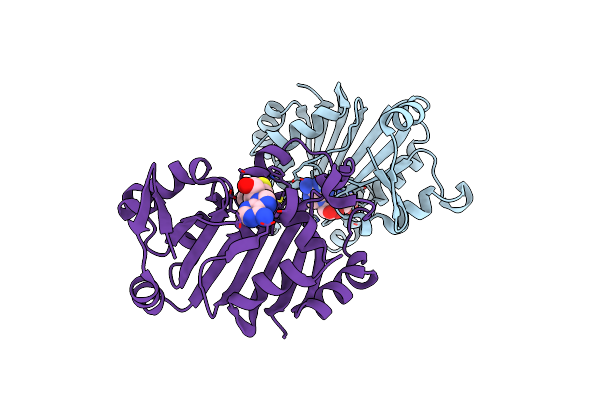
A Novel High Affinity Integrin Alphaiibbeta3 Receptor Antagonist That Unexpectedly Displaces Mg2+ From The Beta3 Midas
Organism: Homo sapiens, Mus musculus
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2012-03-28 Classification: CELL ADHESION Ligands: CA, RC2, SO4, NAG, CL |
 |
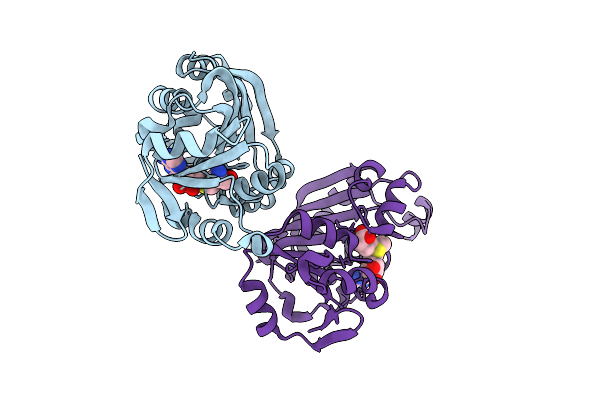
A Novel High Affinity Integrin Alphaiibbeta3 Receptor Antagonist That Unexpectedly Displaces Mg2+ From The Beta3 Midas
Organism: Homo sapiens, Mus musculus
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2012-03-28 Classification: CELL ADHESION Ligands: SO4, GOL, CA, CL, MG, NAG |
 |
Organism: Streptomyces sp. dsm 40477
Method: X-RAY DIFFRACTION Resolution:1.69 Å Release Date: 2010-12-08 Classification: TRANSFERASE Ligands: SAH |
 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.81 Å Release Date: 2010-12-08 Classification: TRANSFERASE Ligands: SAM |
 |
Structure Of Klenow Fragment Of Taq Polymerase In Complex With An Abasic Site
Organism: Thermus aquaticus
Method: X-RAY DIFFRACTION Resolution:2.25 Å Release Date: 2010-05-05 Classification: TRANSFERASE/DNA Ligands: DDS, MG, ACT, GOL, NA |
 |
Structure Of The Large Fragment Of Thermus Aquaticus Dna Polymerase I In Complex With A Blunt-Ended Dna And Ddatp
Organism: Thermus aquaticus
Method: X-RAY DIFFRACTION Resolution:2.19 Å Release Date: 2010-05-05 Classification: TRANSFERASE/DNA Ligands: DDS, ACT, GOL, MG, NA |
 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:3.10 Å Release Date: 2007-04-24 Classification: HYDROLASE/DNA |
 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.20 Å Release Date: 2007-04-24 Classification: HYDROLASE |
 |
Organism: Equus caballus
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2005-07-26 Classification: LIPID TRANSPORT Ligands: EPE, LDA |
 |
Crystal Structure Of Connective Tissue Activating Peptide-Iii(Ctap-Iii) Complexed With Polyvinylsulfonic Acid
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.93 Å Release Date: 2003-08-26 Classification: BLOOD CLOTTING Ligands: ESA |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2003-08-26 Classification: BLOOD CLOTTING |

